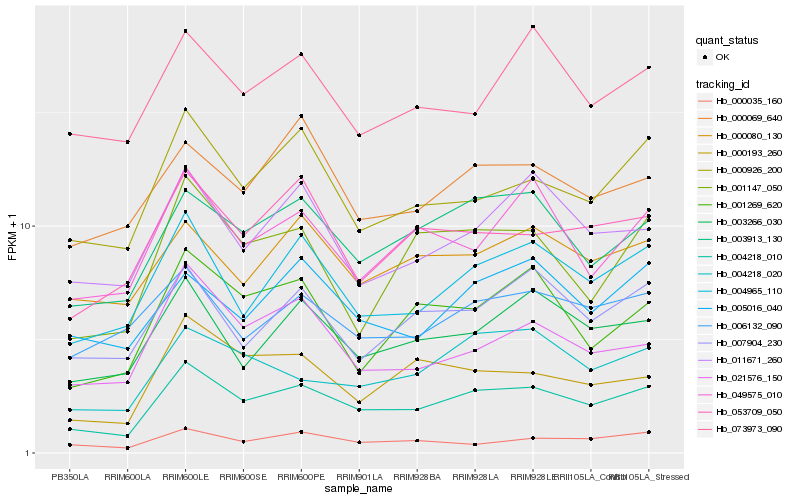
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 2 |
Hb_003913_130 |
0.0874814979 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 3 |
Hb_004965_110 |
0.0892255126 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 4 |
Hb_000926_200 |
0.098340218 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_003266_030 |
0.1020473788 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 6 |
Hb_053709_050 |
0.1069101807 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_004218_020 |
0.1093985193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000080_130 |
0.110298583 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 9 |
Hb_007904_230 |
0.1103735176 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000035_160 |
0.1106767074 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 11 |
Hb_011671_260 |
0.1106885814 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_049575_010 |
0.1109668238 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 13 |
Hb_000193_260 |
0.1113136648 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 14 |
Hb_001147_050 |
0.1118899136 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 15 |
Hb_021576_150 |
0.1119329696 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 16 |
Hb_001269_620 |
0.1132770234 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 17 |
Hb_005016_040 |
0.1137203753 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 18 |
Hb_006132_090 |
0.1153463493 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000069_640 |
0.1162649047 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 20 |
Hb_073973_090 |
0.1172483989 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |