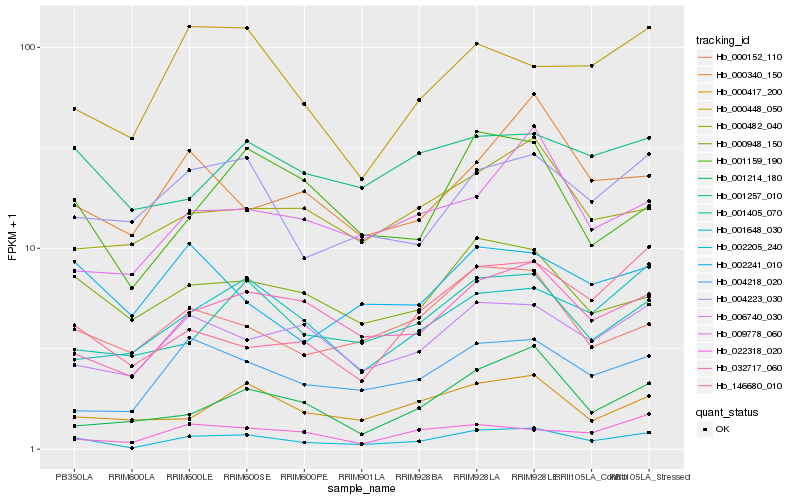
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_240 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa005987mg [Prunus persica] |
| 2 |
Hb_032717_060 |
0.1421419933 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004218_020 |
0.1544887424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006740_030 |
0.1549243341 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_001159_190 |
0.1552881897 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_001405_070 |
0.1560456223 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 7 |
Hb_004223_030 |
0.1573416077 |
- |
- |
PREDICTED: uncharacterized protein LOC105635347 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000152_110 |
0.1573483731 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_000948_150 |
0.1578243037 |
- |
- |
PREDICTED: heme-binding-like protein At3g10130, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000417_200 |
0.1656290502 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001648_030 |
0.1684564729 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000448_050 |
0.1693717633 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_146680_010 |
0.1716133047 |
- |
- |
hypothetical protein JCGZ_15486 [Jatropha curcas] |
| 14 |
Hb_002241_010 |
0.1747241308 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 15 |
Hb_022318_020 |
0.1757705941 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 16 |
Hb_000340_150 |
0.1764489678 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 17 |
Hb_009778_060 |
0.1771993888 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 18 |
Hb_000482_040 |
0.1776737831 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 19 |
Hb_001214_180 |
0.1783523269 |
- |
- |
- |
| 20 |
Hb_001257_010 |
0.1783884891 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |