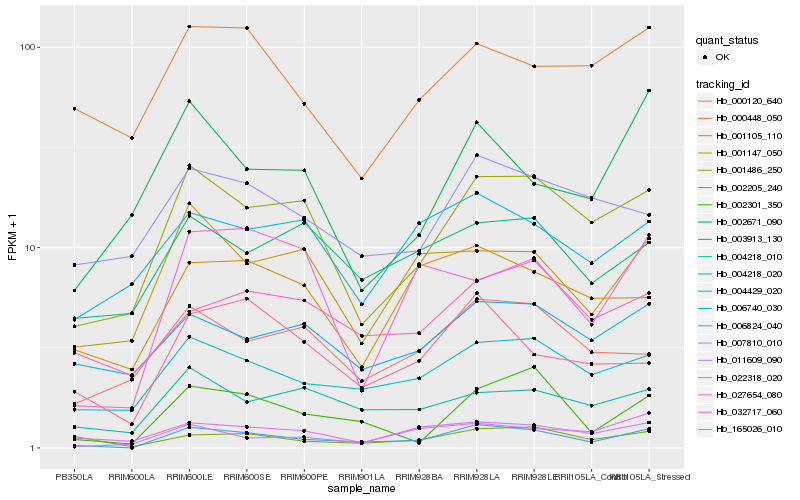
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006824_040 |
0.0 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Populus euphratica] |
| 2 |
Hb_004218_020 |
0.1691793537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001486_250 |
0.1697390474 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 4 |
Hb_002301_350 |
0.1731582283 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 5 |
Hb_002205_240 |
0.1862910087 |
- |
- |
hypothetical protein PRUPE_ppa005987mg [Prunus persica] |
| 6 |
Hb_011609_090 |
0.1942102451 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_004218_010 |
0.1972429872 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 8 |
Hb_165026_010 |
0.1972730805 |
transcription factor |
TF Family: NAC |
hypothetical protein JCGZ_14166 [Jatropha curcas] |
| 9 |
Hb_001147_050 |
0.2028517593 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 10 |
Hb_022318_020 |
0.2063514544 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 11 |
Hb_032717_060 |
0.2082958038 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006740_030 |
0.2093853796 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000120_640 |
0.2118910542 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001105_110 |
0.2124775281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_027654_080 |
0.2128570369 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003913_130 |
0.2140285256 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 17 |
Hb_007810_010 |
0.2156174177 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004429_020 |
0.2181662664 |
- |
- |
PREDICTED: magnesium transporter MRS2-5 [Jatropha curcas] |
| 19 |
Hb_000448_050 |
0.2194186508 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002671_090 |
0.2208616183 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |