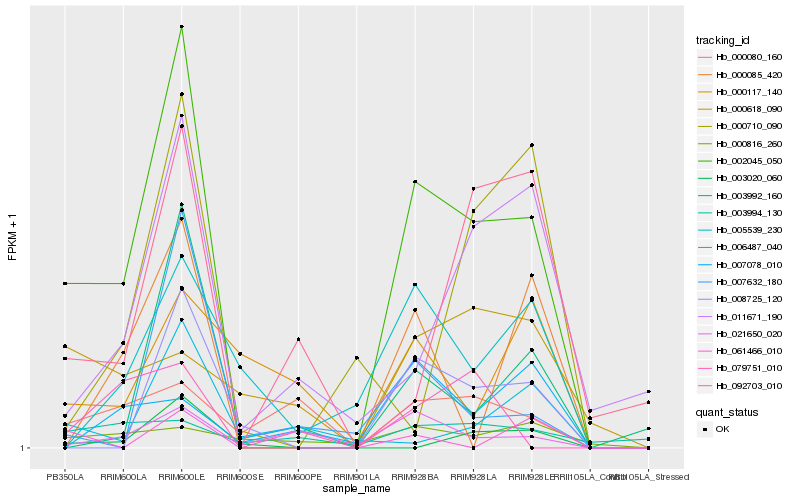
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_008725_120 |
0.3388408233 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Populus euphratica] |
| 3 |
Hb_011671_190 |
0.3419205927 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 4 |
Hb_021650_020 |
0.3470152557 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 5 |
Hb_003020_060 |
0.3521164822 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein [Jatropha curcas] |
| 6 |
Hb_092703_010 |
0.3521891573 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 7 |
Hb_000080_160 |
0.3535556372 |
- |
- |
Peroxin 13 [Theobroma cacao] |
| 8 |
Hb_003992_160 |
0.3594788122 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 9 |
Hb_000085_420 |
0.3605672092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000618_090 |
0.3612419176 |
- |
- |
- |
| 11 |
Hb_000816_260 |
0.3640438962 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_003994_130 |
0.3677510874 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 13 |
Hb_005539_230 |
0.3683426254 |
- |
- |
hypothetical protein L484_027799 [Morus notabilis] |
| 14 |
Hb_007632_180 |
0.3773523165 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 15 |
Hb_079751_010 |
0.3804476615 |
- |
- |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000710_090 |
0.3832139247 |
- |
- |
PREDICTED: uncharacterized protein LOC104583249 [Brachypodium distachyon] |
| 17 |
Hb_061466_010 |
0.3844159169 |
- |
- |
hypothetical protein POPTR_0013s14640g [Populus trichocarpa] |
| 18 |
Hb_007078_010 |
0.3884316595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006487_040 |
0.3925254249 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 20 |
Hb_000117_140 |
0.3938106319 |
- |
- |
hypothetical protein CISIN_1g042884mg, partial [Citrus sinensis] |