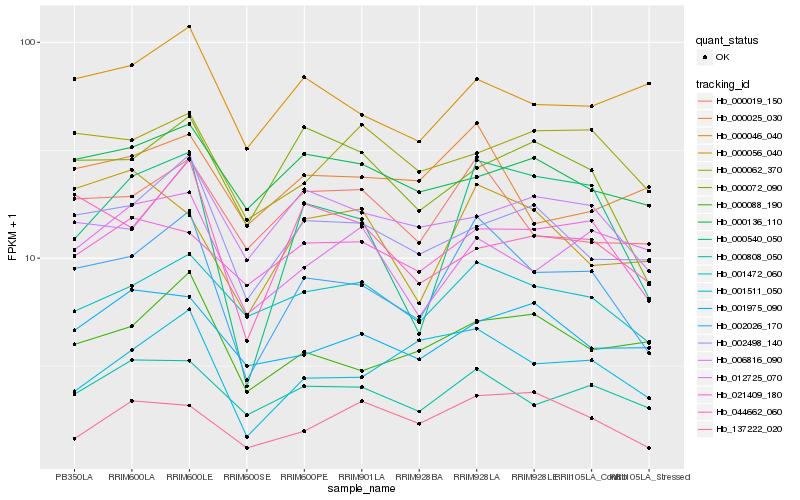
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002026_170 |
0.0 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 2 |
Hb_000540_050 |
0.1181902392 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 3 |
Hb_000019_150 |
0.1319518909 |
- |
- |
PREDICTED: uncharacterized protein LOC105630208 [Jatropha curcas] |
| 4 |
Hb_001472_060 |
0.1334069731 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000808_050 |
0.134331879 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 6 |
Hb_001511_050 |
0.1355797181 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase bub1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002498_140 |
0.1404509867 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 8 |
Hb_000088_190 |
0.1494516696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000072_090 |
0.1520005652 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 10 |
Hb_044662_060 |
0.1533935135 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000025_030 |
0.1560818999 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_021409_180 |
0.1579495504 |
- |
- |
PREDICTED: glutamine-dependent NAD(+) synthetase [Jatropha curcas] |
| 13 |
Hb_006816_090 |
0.1585536368 |
- |
- |
PREDICTED: uncharacterized protein LOC105647425 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000046_040 |
0.1590785531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012725_070 |
0.1622464116 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000062_370 |
0.1626377818 |
- |
- |
beta-ketoacyl-ACP reductase [Vernicia fordii] |
| 17 |
Hb_000056_040 |
0.1639963807 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 18 |
Hb_137222_020 |
0.1642182746 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000136_110 |
0.1650632631 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 20 |
Hb_001975_090 |
0.1673327318 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |