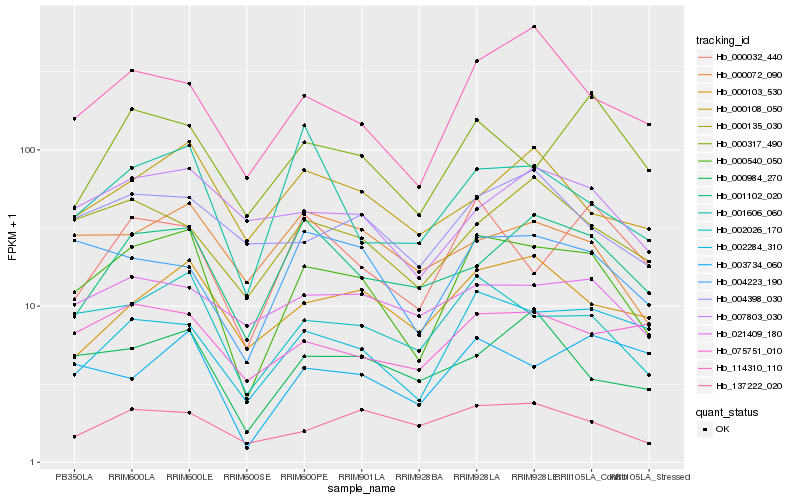
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000540_050 |
0.0 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 2 |
Hb_002026_170 |
0.1181902392 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 3 |
Hb_002284_310 |
0.1561334813 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 4 |
Hb_114310_110 |
0.1653571222 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_004223_190 |
0.1693105094 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 6 |
Hb_000032_440 |
0.1746639073 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_000103_530 |
0.1750097016 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 8 |
Hb_000135_030 |
0.1756300031 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007803_030 |
0.17632553 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000984_270 |
0.1767030232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_137222_020 |
0.1786142222 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001102_020 |
0.17893056 |
- |
- |
solanesyl diphosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_000108_050 |
0.1812755509 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 14 |
Hb_000317_490 |
0.1814053479 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 15 |
Hb_003734_060 |
0.1820114186 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000072_090 |
0.1828514696 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 17 |
Hb_004398_030 |
0.1847308247 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 18 |
Hb_075751_010 |
0.1855227864 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001606_060 |
0.1860382385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_021409_180 |
0.1863541897 |
- |
- |
PREDICTED: glutamine-dependent NAD(+) synthetase [Jatropha curcas] |