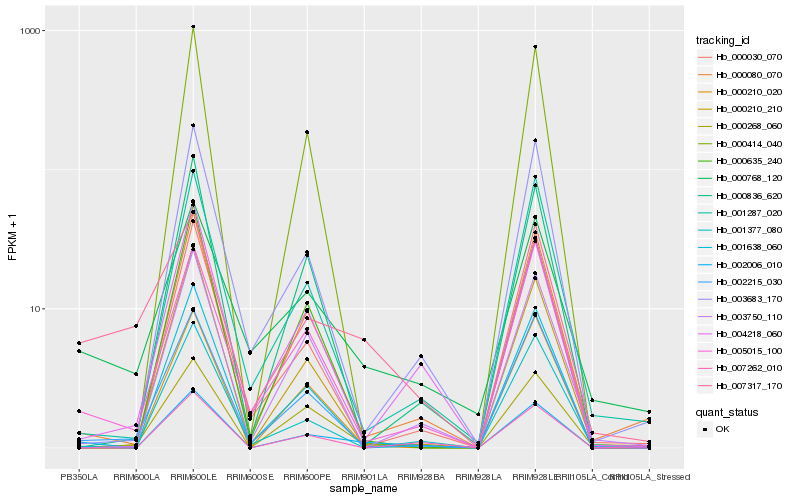
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002006_010 |
0.0 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_000080_070 |
0.1649061381 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005015_100 |
0.1752765749 |
- |
- |
PREDICTED: uncharacterized protein LOC105636541 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000268_060 |
0.1767231497 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_002215_030 |
0.1771400976 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007262_010 |
0.1845237946 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000836_620 |
0.1845322715 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001377_080 |
0.1857593202 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 9 |
Hb_004218_060 |
0.1862745041 |
- |
- |
annexin [Manihot esculenta] |
| 10 |
Hb_001638_060 |
0.1888661972 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 11 |
Hb_001287_020 |
0.1893410828 |
- |
- |
SulA [Manihot esculenta] |
| 12 |
Hb_003683_170 |
0.189551193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000768_120 |
0.1916292576 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 14 |
Hb_003750_110 |
0.1920662641 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 15 |
Hb_000210_210 |
0.1925965477 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000210_020 |
0.1929731077 |
transcription factor |
TF Family: OFP |
Ovate family protein 13, putative [Theobroma cacao] |
| 17 |
Hb_000030_070 |
0.1933296448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000635_240 |
0.1944019124 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 19 |
Hb_007317_170 |
0.194424616 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 20 |
Hb_000414_040 |
0.1949298078 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |