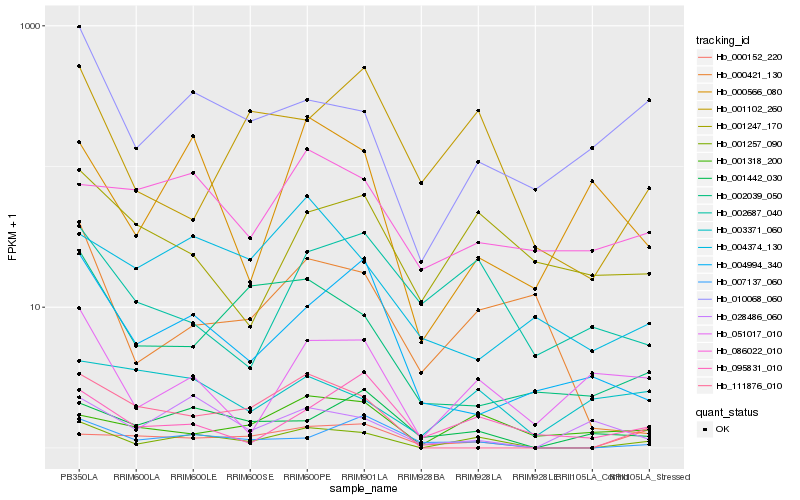
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001257_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 2 |
Hb_051017_010 |
0.2743176503 |
- |
- |
Uncharacterized protein TCM_031047 [Theobroma cacao] |
| 3 |
Hb_010068_060 |
0.2757349263 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 4 |
Hb_111876_010 |
0.2824173419 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 5 |
Hb_007137_060 |
0.2828460959 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 6 |
Hb_000152_220 |
0.2911976565 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 7 |
Hb_001442_030 |
0.2983540488 |
- |
- |
Peroxisome biogenesis factor [Medicago truncatula] |
| 8 |
Hb_001318_200 |
0.3083059682 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 9 |
Hb_001102_260 |
0.3085274108 |
- |
- |
hypothetical protein L484_019841 [Morus notabilis] |
| 10 |
Hb_002039_050 |
0.3099789531 |
- |
- |
PREDICTED: U-box domain-containing protein 40 [Jatropha curcas] |
| 11 |
Hb_028486_060 |
0.3127153512 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 12 |
Hb_095831_010 |
0.314873184 |
- |
- |
PREDICTED: uncharacterized protein LOC104884649 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_000566_080 |
0.3161597644 |
- |
- |
- |
| 14 |
Hb_000421_130 |
0.3165832091 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_001247_170 |
0.3182949054 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 16 |
Hb_086022_010 |
0.3188715059 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 17 |
Hb_004994_340 |
0.3191203369 |
- |
- |
- |
| 18 |
Hb_003371_060 |
0.320660601 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase, mitochondrial isoform X3 [Populus euphratica] |
| 19 |
Hb_004374_130 |
0.3208706377 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 20 |
Hb_002687_040 |
0.3217399428 |
- |
- |
PREDICTED: alpha-galactosidase 1-like [Jatropha curcas] |