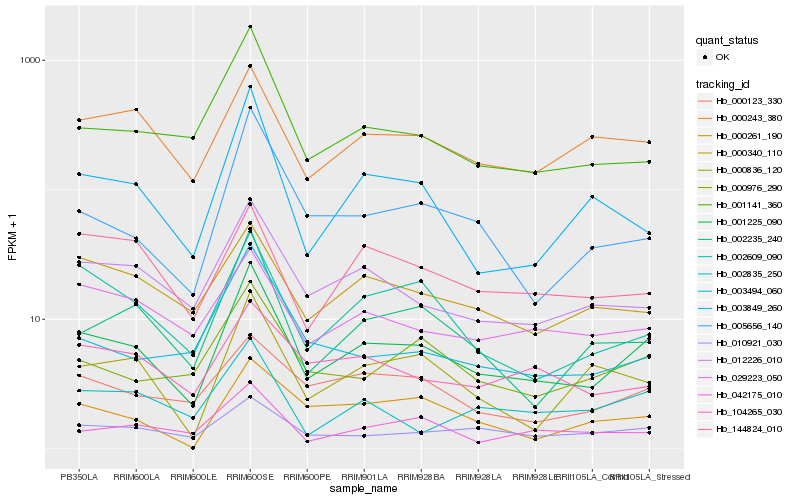
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001225_090 |
0.0 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_012226_010 |
0.1425791719 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 3 |
Hb_002235_240 |
0.1481354771 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 4 |
Hb_001141_360 |
0.1483199902 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 5 |
Hb_005656_140 |
0.1508836889 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 6 |
Hb_000976_290 |
0.1520349218 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 7 |
Hb_002609_090 |
0.1554009069 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000243_380 |
0.1561881521 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 9 |
Hb_003849_260 |
0.1591597885 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 10 |
Hb_010921_030 |
0.1591855589 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At2g01810 [Jatropha curcas] |
| 11 |
Hb_000836_120 |
0.1612435041 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 12 |
Hb_003494_060 |
0.163137064 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 13 |
Hb_029223_050 |
0.1651639187 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000123_330 |
0.1665902007 |
- |
- |
hypothetical protein M569_14136, partial [Genlisea aurea] |
| 15 |
Hb_000340_110 |
0.1672395289 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 16 |
Hb_042175_010 |
0.1711135192 |
- |
- |
hypothetical protein CISIN_1g0016721mg, partial [Citrus sinensis] |
| 17 |
Hb_104265_030 |
0.1713534873 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 18 |
Hb_000261_190 |
0.1734386941 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Jatropha curcas] |
| 19 |
Hb_002835_250 |
0.174764038 |
- |
- |
octicosapeptide/Phox/Bem1p domain-containing family protein [Populus trichocarpa] |
| 20 |
Hb_144824_010 |
0.1775326081 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |