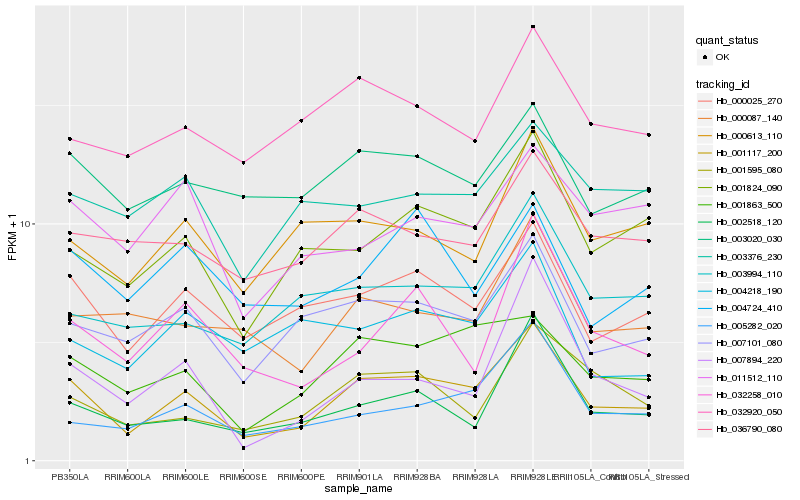
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_200 |
0.0 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000025_270 |
0.1275392446 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_001863_500 |
0.1301001384 |
- |
- |
Protein brittle-1, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_001824_090 |
0.1319569849 |
- |
- |
- |
| 5 |
Hb_007101_080 |
0.1330755464 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 6 |
Hb_005282_020 |
0.1456574651 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |
| 7 |
Hb_002518_120 |
0.151649546 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |
| 8 |
Hb_003994_110 |
0.1519182607 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 9 |
Hb_001595_080 |
0.1526394389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_032258_010 |
0.1535061642 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 11 |
Hb_000613_110 |
0.1544817612 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 12 |
Hb_003020_030 |
0.1561841577 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_000087_140 |
0.1562008728 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 14 |
Hb_036790_080 |
0.156987184 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 15 |
Hb_007894_220 |
0.1591820411 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 16 |
Hb_032920_050 |
0.1597575592 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_011512_110 |
0.1640699143 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_004218_190 |
0.1652448401 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004724_410 |
0.1675082822 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003376_230 |
0.1694258664 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |