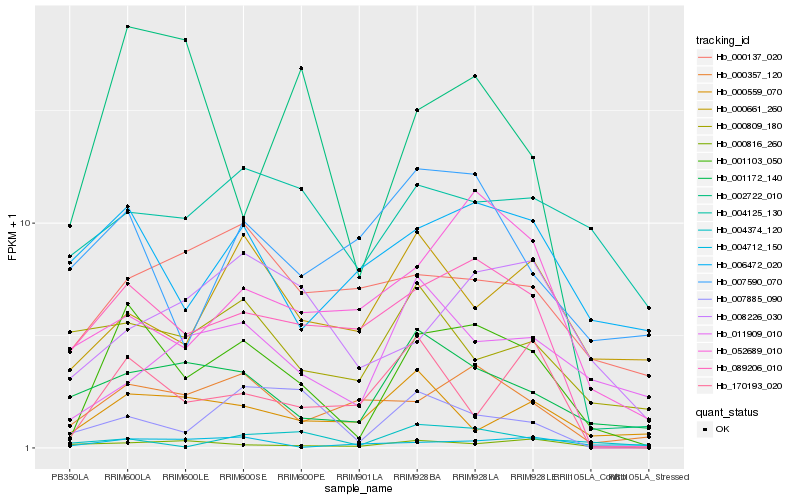
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001103_050 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Nelumbo nucifera] |
| 2 |
Hb_089206_010 |
0.2459654648 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 3 |
Hb_000357_120 |
0.2673205108 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 4 |
Hb_001172_140 |
0.2764665149 |
- |
- |
hypothetical protein POPTR_0014s17790g [Populus trichocarpa] |
| 5 |
Hb_006472_020 |
0.2863096674 |
- |
- |
heat shock protein [Cicer arietinum] |
| 6 |
Hb_008226_030 |
0.2988747979 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 7 |
Hb_170193_020 |
0.3017342119 |
- |
- |
- |
| 8 |
Hb_002722_010 |
0.3036966417 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 9 |
Hb_007885_090 |
0.3037858672 |
- |
- |
hypothetical protein POPTR_0001s08950g [Populus trichocarpa] |
| 10 |
Hb_052689_010 |
0.3043950795 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 11 |
Hb_004125_130 |
0.3063169281 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 12 |
Hb_000816_260 |
0.3064279201 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 13 |
Hb_000137_020 |
0.3066743639 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 14 |
Hb_007590_070 |
0.3097266341 |
- |
- |
PREDICTED: inositol 3-kinase [Jatropha curcas] |
| 15 |
Hb_000661_260 |
0.3104696872 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 16 |
Hb_000559_070 |
0.3107513598 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 17 |
Hb_000809_180 |
0.3119475196 |
- |
- |
- |
| 18 |
Hb_011909_010 |
0.3122884194 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 19 |
Hb_004374_120 |
0.3129703513 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 20 |
Hb_004712_150 |
0.3139539827 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |