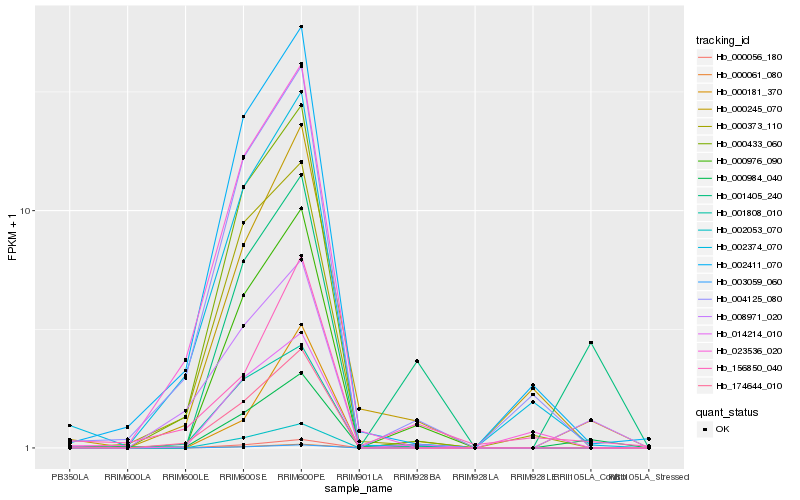
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000984_040 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_001405_240 |
0.1779711303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008971_020 |
0.1828628602 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Eucalyptus grandis] |
| 4 |
Hb_014214_010 |
0.1925547773 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_002411_070 |
0.2004500237 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 6 |
Hb_000433_060 |
0.203757035 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 7 |
Hb_000976_090 |
0.2039732085 |
- |
- |
- |
| 8 |
Hb_004125_080 |
0.2048538968 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 9 |
Hb_002053_070 |
0.2058806652 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 14 [Populus euphratica] |
| 10 |
Hb_023536_020 |
0.2085240223 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 11 |
Hb_002374_070 |
0.2094946003 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_000373_110 |
0.2095429003 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 13 |
Hb_001808_010 |
0.2103591101 |
- |
- |
PREDICTED: dirigent protein 23-like [Jatropha curcas] |
| 14 |
Hb_174644_010 |
0.2154411727 |
- |
- |
- |
| 15 |
Hb_000181_370 |
0.2157417853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000056_180 |
0.2163047898 |
- |
- |
- |
| 17 |
Hb_000061_080 |
0.2165072815 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 18 |
Hb_003059_060 |
0.2169207472 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 19 |
Hb_000245_070 |
0.2172354902 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 20 |
Hb_156850_040 |
0.2174755879 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |