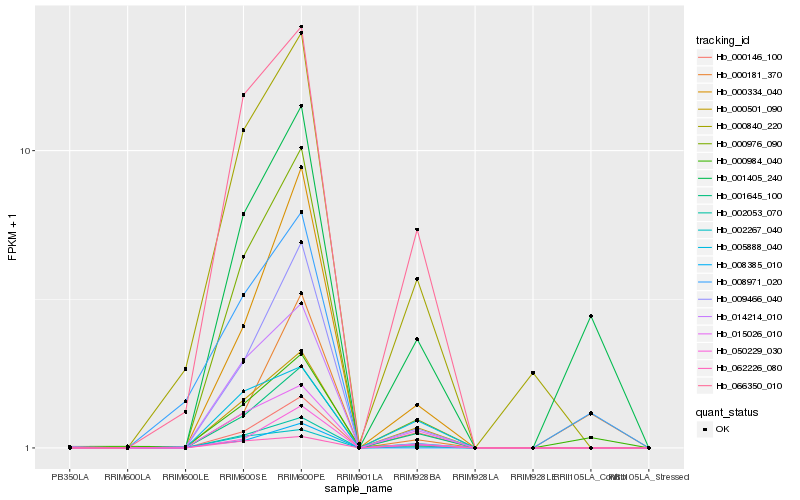
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000984_040 |
0.1779711303 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_000181_370 |
0.1844398468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000501_090 |
0.212341238 |
- |
- |
- |
| 5 |
Hb_001645_100 |
0.2136003282 |
- |
- |
- |
| 6 |
Hb_002053_070 |
0.2258913603 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 14 [Populus euphratica] |
| 7 |
Hb_015026_010 |
0.2305837971 |
- |
- |
- |
| 8 |
Hb_000976_090 |
0.2311585293 |
- |
- |
- |
| 9 |
Hb_066350_010 |
0.232302882 |
- |
- |
somatic embryogenesis receptor-like kinase 4, partial [Rosa hybrid cultivar] |
| 10 |
Hb_009466_040 |
0.2331252933 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 11 |
Hb_002267_040 |
0.2362360984 |
- |
- |
- |
| 12 |
Hb_000334_040 |
0.2397511178 |
- |
- |
hypothetical protein POPTR_0001s41860g [Populus trichocarpa] |
| 13 |
Hb_050229_030 |
0.2428993076 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_062226_080 |
0.2435215256 |
- |
- |
- |
| 15 |
Hb_008385_010 |
0.2442025139 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 16 |
Hb_005888_040 |
0.2465631929 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Jatropha curcas] |
| 17 |
Hb_000146_100 |
0.2519049298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_014214_010 |
0.2527612544 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000840_220 |
0.2530559369 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 20 |
Hb_008971_020 |
0.2543683661 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Eucalyptus grandis] |