| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
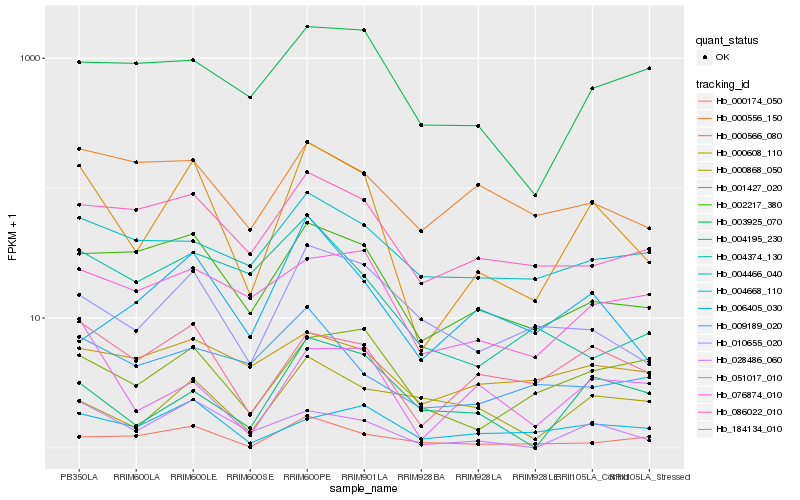
Hb_000566_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_028486_060 |
0.1833021202 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 3 |
Hb_184134_010 |
0.1909121219 |
- |
- |
PREDICTED: variable charge X-linked protein 3B [Jatropha curcas] |
| 4 |
Hb_002217_380 |
0.2013430665 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 5 |
Hb_010655_020 |
0.2115469298 |
- |
- |
hypothetical protein CICLE_v10032410mg [Citrus clementina] |
| 6 |
Hb_086022_010 |
0.2257624551 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 7 |
Hb_001427_020 |
0.2269254925 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 8 |
Hb_004668_110 |
0.2307762586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_076874_010 |
0.2356676942 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 10 |
Hb_051017_010 |
0.236943304 |
- |
- |
Uncharacterized protein TCM_031047 [Theobroma cacao] |
| 11 |
Hb_000174_050 |
0.2371731771 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 [Populus euphratica] |
| 12 |
Hb_004195_230 |
0.2394111603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003925_070 |
0.2507334216 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 14 |
Hb_000868_050 |
0.2528243244 |
- |
- |
RAD51 homolog [Populus nigra] |
| 15 |
Hb_009189_020 |
0.2540275661 |
- |
- |
PREDICTED: CASP-like protein 4B4 [Jatropha curcas] |
| 16 |
Hb_004466_040 |
0.2556612679 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 17 |
Hb_000556_150 |
0.2561662354 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 18 |
Hb_004374_130 |
0.2566122327 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 19 |
Hb_000608_110 |
0.2577595028 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 20 |
Hb_006405_030 |
0.2581111278 |
- |
- |
hypothetical protein VITISV_043238 [Vitis vinifera] |