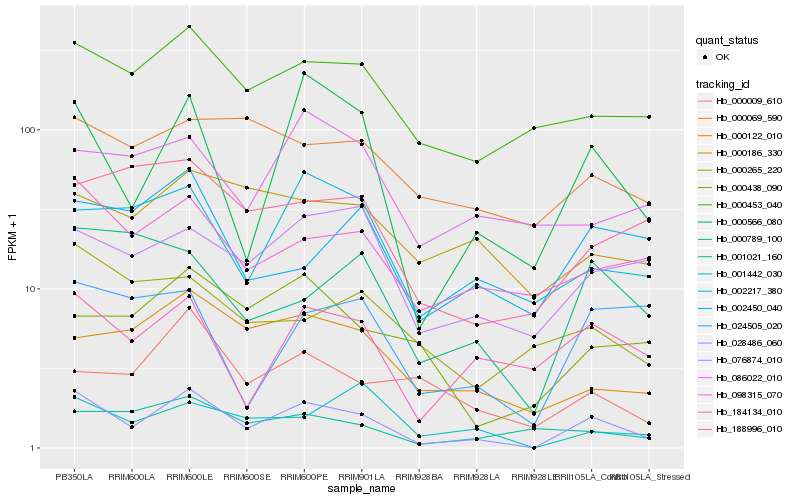
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028486_060 |
0.0 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 2 |
Hb_000566_080 |
0.1833021202 |
- |
- |
- |
| 3 |
Hb_184134_010 |
0.2193929983 |
- |
- |
PREDICTED: variable charge X-linked protein 3B [Jatropha curcas] |
| 4 |
Hb_000453_040 |
0.2222781822 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 5 |
Hb_098315_070 |
0.2227972786 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002217_380 |
0.2308743259 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 7 |
Hb_076874_010 |
0.237328826 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 8 |
Hb_000122_010 |
0.2400039104 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_001442_030 |
0.2406465282 |
- |
- |
Peroxisome biogenesis factor [Medicago truncatula] |
| 10 |
Hb_000789_100 |
0.2428230484 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |
| 11 |
Hb_001021_160 |
0.2484116779 |
- |
- |
PREDICTED: homeobox protein LUMINIDEPENDENS [Jatropha curcas] |
| 12 |
Hb_024505_020 |
0.250397941 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 13 |
Hb_002450_040 |
0.2522346557 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 6B [Vitis vinifera] |
| 14 |
Hb_188996_010 |
0.2546425644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000009_610 |
0.2546741915 |
- |
- |
PREDICTED: uncharacterized protein LOC105639638 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000438_090 |
0.2560490289 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 17 |
Hb_000186_330 |
0.2572075963 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 18 |
Hb_000265_220 |
0.2581573988 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 19 |
Hb_000069_590 |
0.2583600995 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 20 |
Hb_086022_010 |
0.2584078348 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |