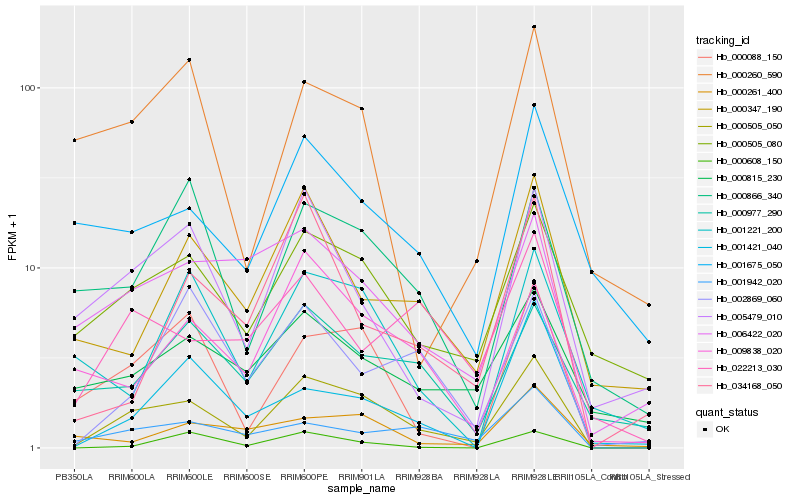
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000505_050 |
0.0 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X2 [Gossypium raimondii] |
| 2 |
Hb_000088_150 |
0.1995626397 |
- |
- |
hypothetical protein POPTR_0013s11250g [Populus trichocarpa] |
| 3 |
Hb_000505_080 |
0.2067039339 |
- |
- |
Inner membrane transport protein yjjL, putative [Ricinus communis] |
| 4 |
Hb_001942_020 |
0.2205538295 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_009838_020 |
0.2300123643 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 6 |
Hb_005479_010 |
0.2306256844 |
- |
- |
PREDICTED: uncharacterized protein LOC105111002 [Populus euphratica] |
| 7 |
Hb_000866_340 |
0.2353803791 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_000347_190 |
0.2384104616 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 9 |
Hb_001675_050 |
0.2412333246 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 10 |
Hb_000608_150 |
0.2422695347 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000977_290 |
0.2454197588 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000815_230 |
0.2457138718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_034168_050 |
0.246121532 |
- |
- |
Photosystem I assembly protein Ycf3 [Medicago truncatula] |
| 14 |
Hb_006422_020 |
0.2467067238 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 15 |
Hb_001221_200 |
0.2482591254 |
- |
- |
- |
| 16 |
Hb_022213_030 |
0.2489707748 |
- |
- |
PREDICTED: uncharacterized protein LOC105114631 [Populus euphratica] |
| 17 |
Hb_000261_400 |
0.2516441361 |
- |
- |
hypothetical protein JCGZ_01125 [Jatropha curcas] |
| 18 |
Hb_002869_060 |
0.2524327934 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 19 |
Hb_000260_590 |
0.2531082468 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 20 |
Hb_001421_040 |
0.2533452937 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 2 [Jatropha curcas] |