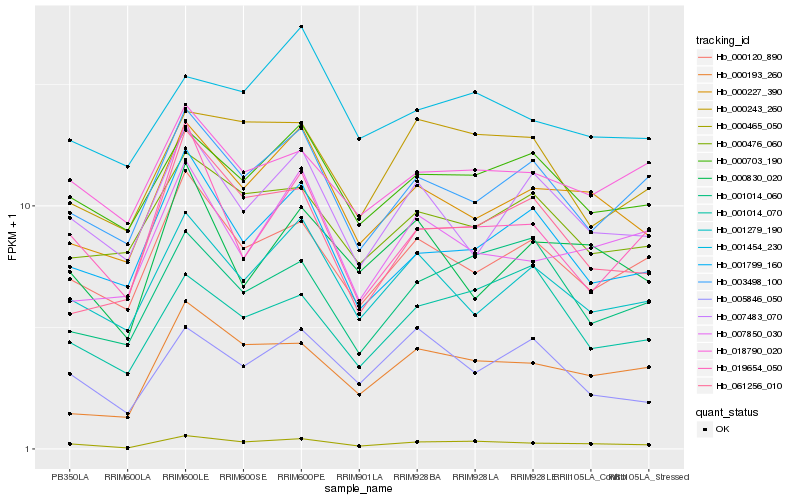
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_050 |
0.0 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH4 [Jatropha curcas] |
| 2 |
Hb_000227_390 |
0.1136412531 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000193_260 |
0.1141545643 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 4 |
Hb_000120_890 |
0.1203697287 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 5 |
Hb_000703_190 |
0.1258339918 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003498_100 |
0.1269155808 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 7 |
Hb_001799_160 |
0.1307811655 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_001279_190 |
0.1312867832 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000243_260 |
0.1322039284 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 10 |
Hb_018790_020 |
0.1327611926 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 11 |
Hb_019654_050 |
0.1328755818 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 12 |
Hb_061256_010 |
0.134482517 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007850_030 |
0.1382282037 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 14 |
Hb_000476_060 |
0.1394186639 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_001014_060 |
0.1408455105 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 16 |
Hb_005846_050 |
0.1409539193 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 17 |
Hb_007483_070 |
0.1414051843 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 18 |
Hb_001454_230 |
0.1415054693 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 19 |
Hb_001014_070 |
0.1419242123 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 20 |
Hb_000830_020 |
0.1424678403 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |