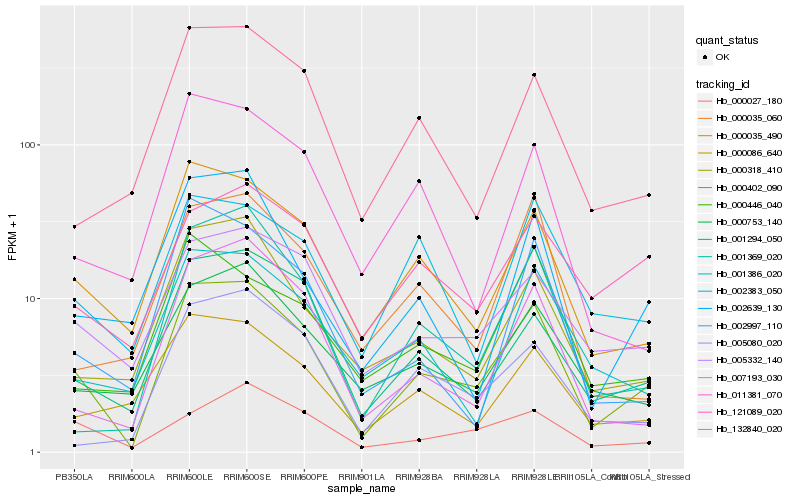
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_090 |
0.0 |
- |
- |
Lipase [Glycine soja] |
| 2 |
Hb_000035_490 |
0.139737816 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 3 |
Hb_001294_050 |
0.1598775548 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 4 |
Hb_005332_140 |
0.1608829915 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000318_410 |
0.1670078077 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 6 |
Hb_000753_140 |
0.1683856222 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 7 |
Hb_000086_640 |
0.1694406942 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_001369_020 |
0.1708514071 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 9 |
Hb_132840_020 |
0.1737946708 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 10 |
Hb_007193_030 |
0.174601549 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002639_130 |
0.1766466453 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 12 |
Hb_001386_020 |
0.1797495337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002383_050 |
0.1805160852 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 14 |
Hb_005080_020 |
0.1851207587 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 15 |
Hb_000027_180 |
0.1853467396 |
- |
- |
PREDICTED: uncharacterized protein LOC105642058 [Jatropha curcas] |
| 16 |
Hb_121089_020 |
0.1857381336 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 17 |
Hb_000446_040 |
0.1877631475 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 18 |
Hb_000035_060 |
0.1881496686 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 19 |
Hb_011381_070 |
0.1892448424 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 20 |
Hb_002997_110 |
0.1895419088 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |