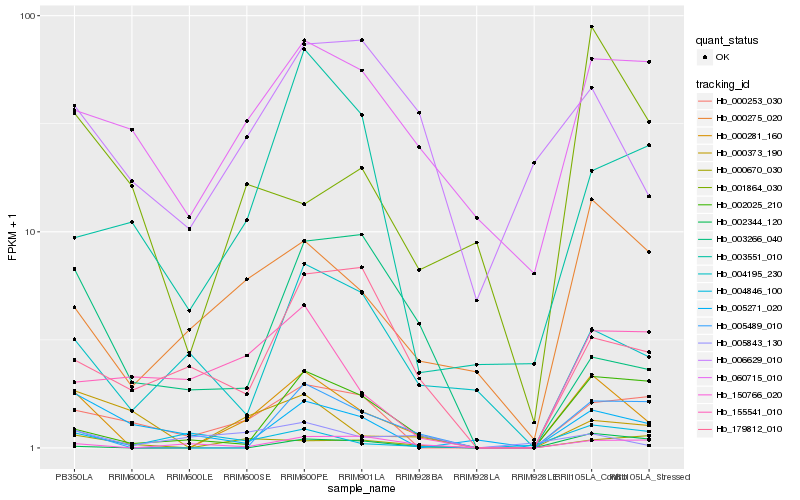
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000281_160 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 716B2-like isoform X2 [Glycine max] |
| 2 |
Hb_000275_020 |
0.2993728784 |
- |
- |
PREDICTED: uncharacterized protein LOC105632656 [Jatropha curcas] |
| 3 |
Hb_005489_010 |
0.3029679477 |
- |
- |
hypothetical protein POPTR_0011s11420g [Populus trichocarpa] |
| 4 |
Hb_002025_210 |
0.3187816248 |
- |
- |
- |
| 5 |
Hb_000670_030 |
0.3221603775 |
- |
- |
- |
| 6 |
Hb_000253_030 |
0.3294234894 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 7 |
Hb_060715_010 |
0.3350730848 |
- |
- |
PREDICTED: notchless protein homolog [Gossypium raimondii] |
| 8 |
Hb_150766_020 |
0.335618201 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 9 |
Hb_002344_120 |
0.3406988163 |
- |
- |
- |
| 10 |
Hb_000373_190 |
0.3433291052 |
- |
- |
- |
| 11 |
Hb_004195_230 |
0.3530065747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006629_010 |
0.3554398827 |
- |
- |
Glutathione S-transferase 2 isoform 3 [Theobroma cacao] |
| 13 |
Hb_005843_130 |
0.3585122148 |
- |
- |
hypothetical protein JCGZ_12389 [Jatropha curcas] |
| 14 |
Hb_004846_100 |
0.3590964172 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 15 |
Hb_179812_010 |
0.3618899674 |
- |
- |
TdcA1-ORF1-ORF2 [Daucus carota] |
| 16 |
Hb_003551_010 |
0.3621359962 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 17 |
Hb_155541_010 |
0.3631444717 |
- |
- |
- |
| 18 |
Hb_005271_020 |
0.3650093428 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 19 |
Hb_001864_030 |
0.365091946 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 20 |
Hb_003266_040 |
0.3681390118 |
- |
- |
PREDICTED: 6.7 kDa chloroplast outer envelope membrane protein-like [Jatropha curcas] |