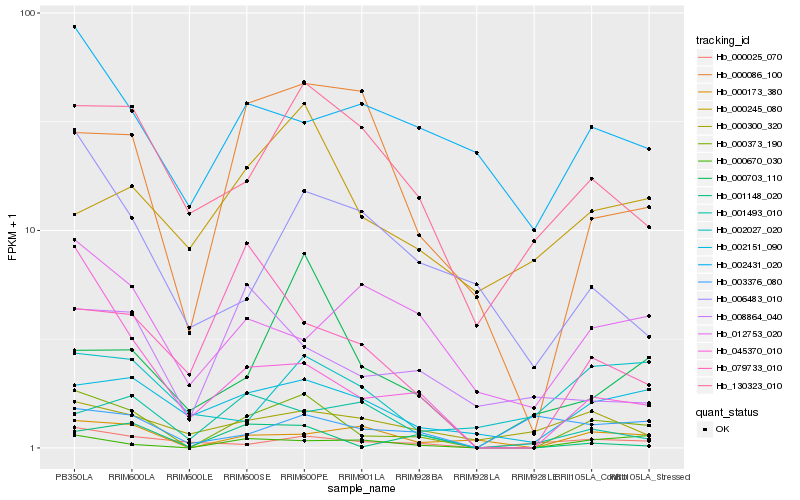
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000373_190 |
0.0 |
- |
- |
- |
| 2 |
Hb_000670_030 |
0.260340885 |
- |
- |
- |
| 3 |
Hb_002151_090 |
0.2742876494 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_045370_010 |
0.2761168132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000173_380 |
0.280861527 |
transcription factor |
TF Family: FAR1 |
- |
| 6 |
Hb_000086_100 |
0.2897023671 |
- |
- |
hypothetical protein POPTR_0005s24730g [Populus trichocarpa] |
| 7 |
Hb_130323_010 |
0.2916304899 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 8 |
Hb_001148_020 |
0.2918348934 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 9 |
Hb_000025_070 |
0.2960790634 |
- |
- |
Uncharacterized protein TCM_040985 [Theobroma cacao] |
| 10 |
Hb_000703_110 |
0.2970381991 |
- |
- |
- |
| 11 |
Hb_000300_320 |
0.3080743268 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 12 |
Hb_002027_020 |
0.3145156047 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 13 |
Hb_079733_010 |
0.3152983862 |
- |
- |
- |
| 14 |
Hb_012753_020 |
0.321768218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003376_080 |
0.3253007065 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000245_080 |
0.3260089542 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 17 |
Hb_001493_010 |
0.3262655151 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 18 |
Hb_006483_010 |
0.327206413 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_002431_020 |
0.3278706915 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 20 |
Hb_008864_040 |
0.3296859305 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |