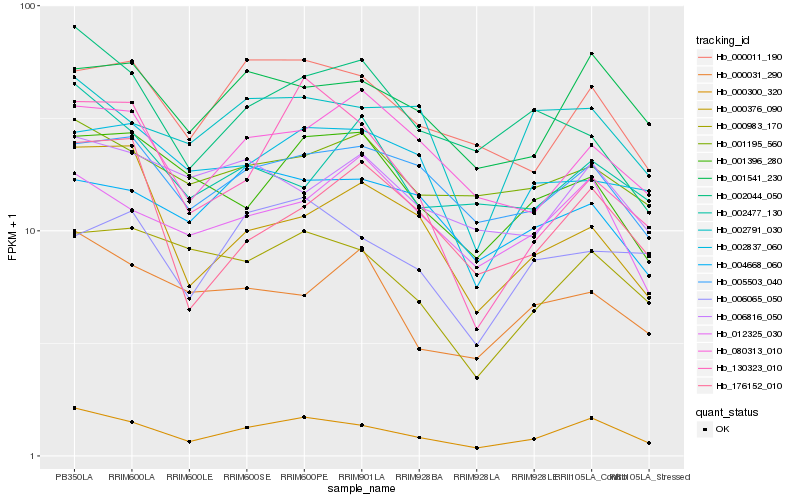
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 2 |
Hb_001541_230 |
0.117741585 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000011_190 |
0.1219395341 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6 [Jatropha curcas] |
| 4 |
Hb_005503_040 |
0.1265965728 |
- |
- |
PREDICTED: AMP deaminase [Jatropha curcas] |
| 5 |
Hb_000983_170 |
0.1269837385 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_012325_030 |
0.135089931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002791_030 |
0.1355058934 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 8 |
Hb_001396_280 |
0.1356004406 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 9 |
Hb_080313_010 |
0.1388768986 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 10 |
Hb_001195_560 |
0.1405385298 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000031_290 |
0.1416334755 |
- |
- |
hypothetical protein POPTR_0006s28540g [Populus trichocarpa] |
| 12 |
Hb_006816_050 |
0.1420509236 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 13 |
Hb_002044_050 |
0.142139763 |
- |
- |
- |
| 14 |
Hb_130323_010 |
0.1423955898 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 15 |
Hb_000376_090 |
0.1425373981 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 16 |
Hb_176152_010 |
0.1447172573 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 17 |
Hb_004668_060 |
0.1455838275 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006065_050 |
0.1492485123 |
transcription factor |
TF Family: AP2 |
PREDICTED: floral homeotic protein APETALA 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002837_060 |
0.1494097629 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 20 |
Hb_002477_130 |
0.1495586235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |