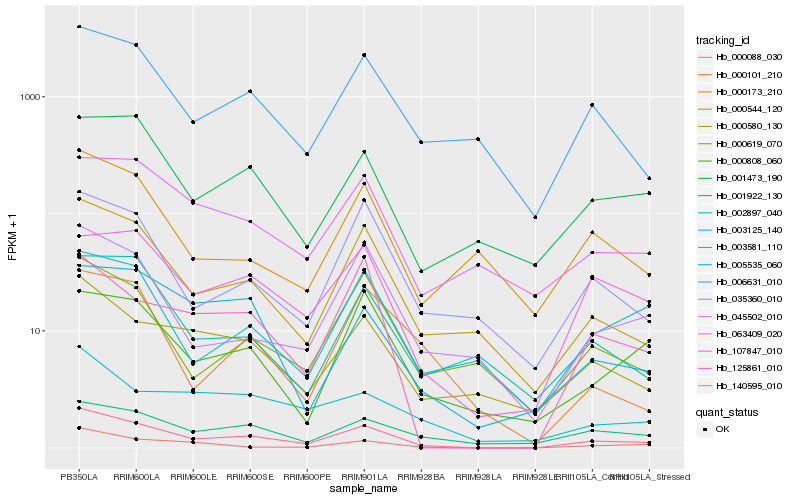
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140595_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000544_120 |
0.1440223754 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 3 |
Hb_000580_130 |
0.1713469559 |
- |
- |
hypothetical protein JCGZ_14541 [Jatropha curcas] |
| 4 |
Hb_006631_010 |
0.1725050405 |
- |
- |
plasma membrane aquaporin 2 [Hevea brasiliensis] |
| 5 |
Hb_107847_010 |
0.1729742687 |
- |
- |
3-isopropylmalate dehydratase, putative [Ricinus communis] |
| 6 |
Hb_125861_010 |
0.1742208805 |
- |
- |
hypothetical protein RCOM_0703970 [Ricinus communis] |
| 7 |
Hb_000088_030 |
0.1787304729 |
- |
- |
hypothetical protein JCGZ_10947 [Jatropha curcas] |
| 8 |
Hb_045502_010 |
0.1789098558 |
- |
- |
prefoldin subunit 1 [Jatropha curcas] |
| 9 |
Hb_001922_130 |
0.1822280235 |
- |
- |
hypothetical protein PHAVU_002G294300g [Phaseolus vulgaris] |
| 10 |
Hb_035360_010 |
0.1830006188 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 11 |
Hb_001473_190 |
0.18531557 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 12 |
Hb_000619_070 |
0.1866344806 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH1 [Jatropha curcas] |
| 13 |
Hb_005535_060 |
0.1905620447 |
- |
- |
- |
| 14 |
Hb_063409_020 |
0.1929059094 |
- |
- |
Calcium-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000808_060 |
0.1942949137 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Jatropha curcas] |
| 16 |
Hb_003581_110 |
0.1956537821 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002897_040 |
0.1971347212 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |
| 18 |
Hb_003125_140 |
0.1976714918 |
- |
- |
PREDICTED: uncharacterized protein LOC105648796 [Jatropha curcas] |
| 19 |
Hb_000173_210 |
0.1982625908 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 20 |
Hb_000101_210 |
0.2016241363 |
- |
- |
PREDICTED: uncharacterized protein LOC105646445 [Jatropha curcas] |