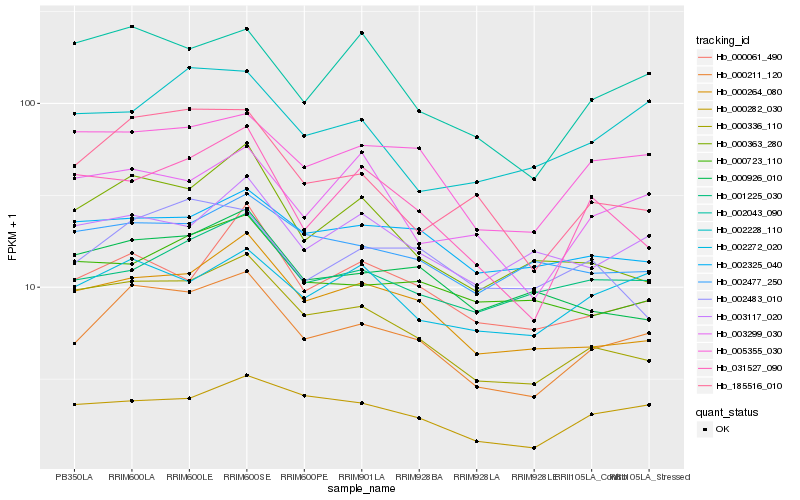
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000211_120 |
0.0 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 2 |
Hb_000336_110 |
0.108167461 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000264_080 |
0.1107483262 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 4 |
Hb_005355_030 |
0.1113751455 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000282_030 |
0.114424353 |
- |
- |
K(+) efflux antiporter 4 [Glycine soja] |
| 6 |
Hb_185516_010 |
0.1144598769 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 7 |
Hb_000061_490 |
0.1175464121 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 8 |
Hb_000363_280 |
0.1213952645 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 9 |
Hb_002272_020 |
0.1218391213 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 10 |
Hb_002043_090 |
0.1232207396 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 11 |
Hb_001225_030 |
0.1265719819 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 12 |
Hb_000926_010 |
0.1296737452 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 13 |
Hb_002477_250 |
0.1325385421 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 14 |
Hb_003117_020 |
0.1326151787 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 15 |
Hb_031527_090 |
0.1329456884 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002325_040 |
0.1331906695 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000723_110 |
0.133952295 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 18 |
Hb_003299_030 |
0.1342083882 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002483_010 |
0.1342115741 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002228_110 |
0.1342629213 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |