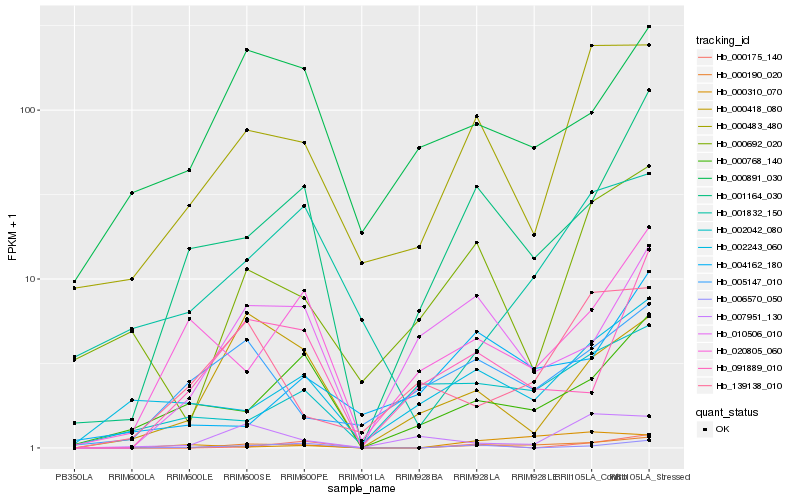
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000190_020 |
0.0 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA [Jatropha curcas] |
| 2 |
Hb_001164_030 |
0.2655473208 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |
| 3 |
Hb_000483_480 |
0.2861892888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010506_010 |
0.3000270482 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 5 |
Hb_000692_020 |
0.3029856571 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL23 [Jatropha curcas] |
| 6 |
Hb_091889_010 |
0.3068506422 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000175_140 |
0.310346509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000768_140 |
0.3105198845 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000418_080 |
0.3137941549 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 10 |
Hb_001832_150 |
0.3164413668 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 11 |
Hb_006570_050 |
0.3167498918 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 12 |
Hb_002243_060 |
0.321615541 |
- |
- |
hypothetical protein POPTR_0022s00750g [Populus trichocarpa] |
| 13 |
Hb_000891_030 |
0.3269276805 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 14 |
Hb_002042_080 |
0.3325585089 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 15 |
Hb_005147_010 |
0.3327001613 |
- |
- |
PREDICTED: uncharacterized protein LOC105629670 [Jatropha curcas] |
| 16 |
Hb_004162_180 |
0.3348151955 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_007951_130 |
0.3357736917 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 18 |
Hb_139138_010 |
0.3360315433 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 19 |
Hb_020805_060 |
0.3395703861 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 20 |
Hb_000310_070 |
0.3415913782 |
- |
- |
BnaA06g08330D [Brassica napus] |