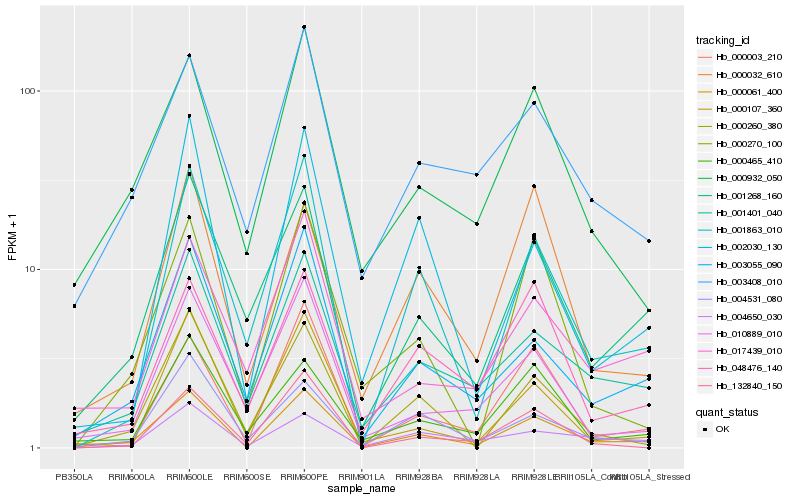
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_400 |
0.0 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 2 |
Hb_001401_040 |
0.1462221629 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 3 |
Hb_000003_210 |
0.1479071685 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 4 |
Hb_003055_090 |
0.154356018 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 5 |
Hb_000932_050 |
0.161813464 |
- |
- |
annexin [Manihot esculenta] |
| 6 |
Hb_001863_010 |
0.1652604063 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 7 |
Hb_001268_160 |
0.1666680673 |
- |
- |
Glycerophosphodiester phosphodiesterase [Medicago truncatula] |
| 8 |
Hb_000260_380 |
0.1686039076 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 9 |
Hb_004531_080 |
0.1744441913 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 10 |
Hb_003408_010 |
0.1771276332 |
- |
- |
fructokinase [Manihot esculenta] |
| 11 |
Hb_010889_010 |
0.1793123269 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 12 |
Hb_000465_410 |
0.1793377541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000270_100 |
0.1797339235 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 14 |
Hb_002030_130 |
0.1800048119 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 15 |
Hb_048476_140 |
0.1829589011 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 16 |
Hb_017439_010 |
0.1838166463 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 17 |
Hb_000107_360 |
0.1855082245 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 18 |
Hb_004650_030 |
0.1880641071 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 19 |
Hb_132840_150 |
0.1882877552 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000032_610 |
0.1893016771 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |