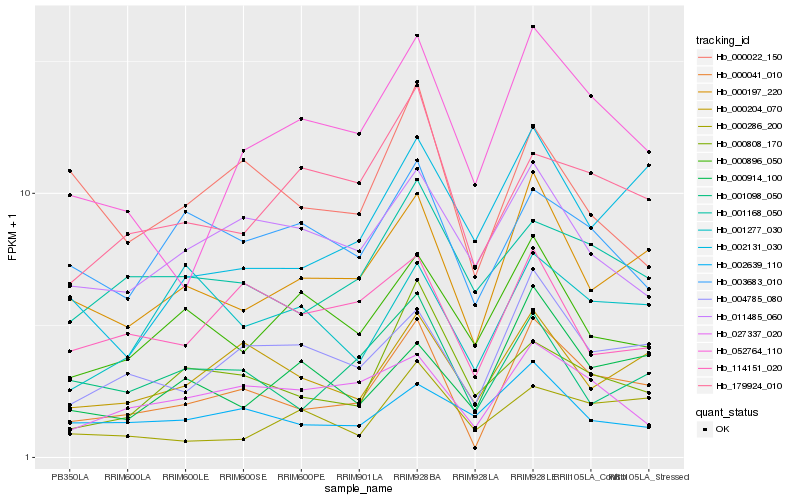
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000041_010 |
0.0 |
- |
- |
pfkB-type carbohydrate kinase family protein [Populus trichocarpa] |
| 2 |
Hb_000197_220 |
0.1376282091 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004785_080 |
0.1405123483 |
- |
- |
hypothetical protein POPTR_0005s21250g [Populus trichocarpa] |
| 4 |
Hb_000204_070 |
0.1546813444 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 5 |
Hb_001098_050 |
0.1697928077 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 6 |
Hb_052764_110 |
0.17049565 |
- |
- |
PREDICTED: quinone oxidoreductase PIG3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_027337_020 |
0.1713130516 |
- |
- |
sodium/hydrogen exchanger plant, putative [Ricinus communis] |
| 8 |
Hb_001168_050 |
0.1815827677 |
- |
- |
hypothetical protein POPTR_0007s03190g [Populus trichocarpa] |
| 9 |
Hb_179924_010 |
0.1818910266 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_000914_100 |
0.1827185514 |
- |
- |
unknown [Glycine max] |
| 11 |
Hb_000808_170 |
0.1853272448 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 12 |
Hb_114151_020 |
0.18805186 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 13 |
Hb_002131_030 |
0.189978612 |
- |
- |
PREDICTED: pyruvate kinase isozyme G, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000022_150 |
0.1936609322 |
- |
- |
- |
| 15 |
Hb_002639_110 |
0.194417385 |
- |
- |
hypothetical protein L484_011673 [Morus notabilis] |
| 16 |
Hb_000896_050 |
0.1965695801 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 17 |
Hb_011485_060 |
0.1965717908 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 18 |
Hb_003683_010 |
0.1967661787 |
- |
- |
longevity assurance factor, putative [Ricinus communis] |
| 19 |
Hb_001277_030 |
0.1970811919 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Nelumbo nucifera] |
| 20 |
Hb_000286_200 |
0.1974657274 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |