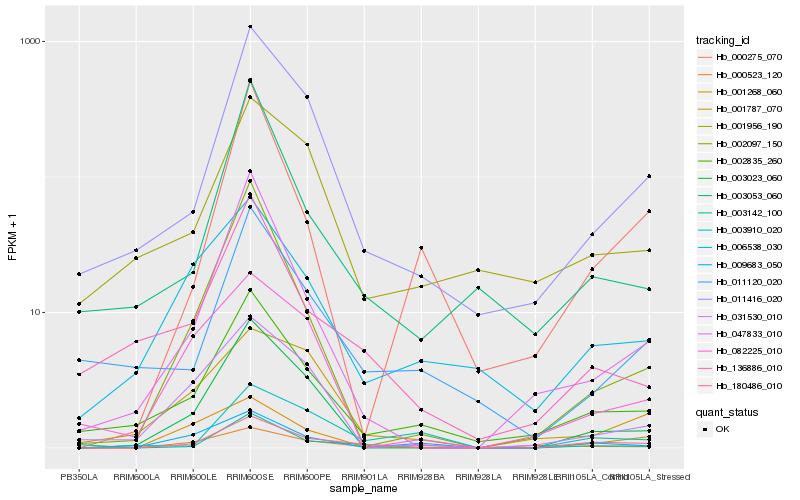
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_180486_010 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_003023_060 |
0.2239755167 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_003142_100 |
0.2324617676 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Eucalyptus grandis] |
| 4 |
Hb_011416_020 |
0.2340967953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001268_060 |
0.2398965001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002835_260 |
0.2411128416 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 7 |
Hb_047833_010 |
0.254880793 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 8 |
Hb_136886_010 |
0.2706884777 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_009683_050 |
0.2732403717 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 10 |
Hb_002097_150 |
0.273987194 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 11 |
Hb_003910_020 |
0.2789500727 |
- |
- |
PREDICTED: reticulon-like protein B13 [Jatropha curcas] |
| 12 |
Hb_006538_030 |
0.2791190147 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 13 |
Hb_003053_060 |
0.2817948534 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 14 |
Hb_001787_070 |
0.2829717606 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Camelina sativa] |
| 15 |
Hb_011120_020 |
0.2840451196 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_031530_010 |
0.2846761121 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 17 |
Hb_001956_190 |
0.2907892639 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 18 |
Hb_082225_010 |
0.291243529 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 19 |
Hb_000523_120 |
0.2937594246 |
- |
- |
PREDICTED: butyrate--CoA ligase AAE11, peroxisomal-like [Jatropha curcas] |
| 20 |
Hb_000275_070 |
0.2940137146 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |