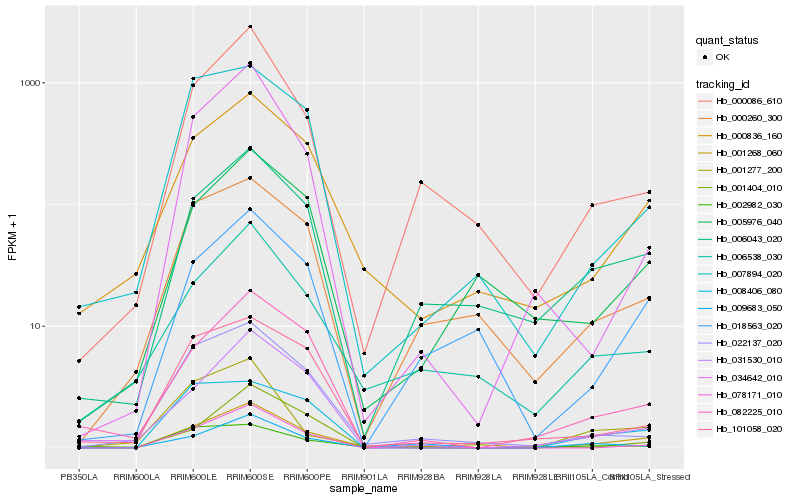
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001268_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000836_160 |
0.16852725 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 3 |
Hb_082225_010 |
0.1808051297 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 4 |
Hb_006043_020 |
0.1816865561 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 5 |
Hb_078171_010 |
0.1825926797 |
- |
- |
PREDICTED: uncharacterized RING finger protein P8B7.15c-like [Jatropha curcas] |
| 6 |
Hb_000086_610 |
0.1862381487 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 7 |
Hb_031530_010 |
0.1874342741 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 8 |
Hb_034642_010 |
0.1949976512 |
- |
- |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 9 |
Hb_009683_050 |
0.1958286812 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 10 |
Hb_018563_020 |
0.1960199134 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 6 [Hevea brasiliensis] |
| 11 |
Hb_008406_080 |
0.1971332858 |
- |
- |
Nonspecific lipid-transfer protein 3 precursor, putative [Ricinus communis] |
| 12 |
Hb_006538_030 |
0.201373058 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 13 |
Hb_022137_020 |
0.2028406534 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 14 |
Hb_005976_040 |
0.2071302912 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 15 |
Hb_000260_300 |
0.210871901 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 16 |
Hb_007894_020 |
0.2120061587 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 17 |
Hb_101058_020 |
0.2132401058 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 18 |
Hb_002982_030 |
0.2143671088 |
- |
- |
Chalcone synthase, putative [Ricinus communis] |
| 19 |
Hb_001277_200 |
0.2193866702 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 20 |
Hb_001404_010 |
0.2201710674 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |