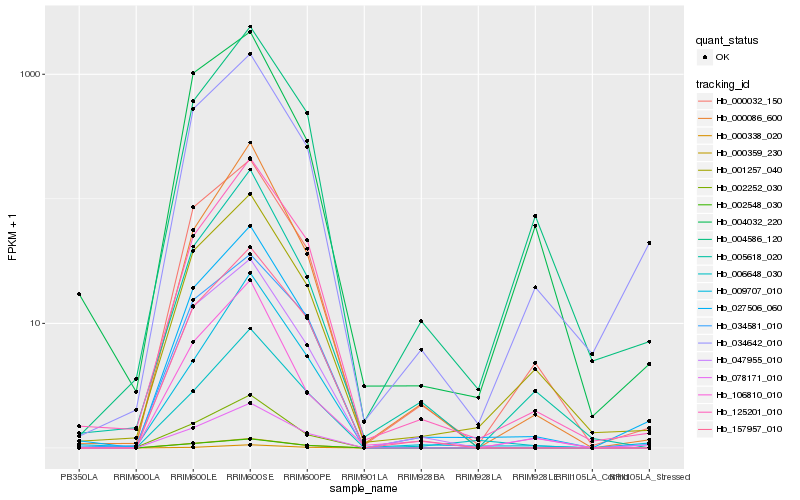
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027506_060 |
0.0 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 2 |
Hb_034642_010 |
0.0801943718 |
- |
- |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 3 |
Hb_125201_010 |
0.0850778812 |
- |
- |
hypothetical protein POPTR_0004s03690g [Populus trichocarpa] |
| 4 |
Hb_047955_010 |
0.089747443 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 5 |
Hb_000032_150 |
0.0912536975 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1691430 [Ricinus communis] |
| 6 |
Hb_001257_040 |
0.0932175322 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 7 |
Hb_000086_600 |
0.1008773992 |
- |
- |
- |
| 8 |
Hb_157957_010 |
0.1009963583 |
- |
- |
hypothetical protein JCGZ_10722 [Jatropha curcas] |
| 9 |
Hb_005618_020 |
0.1015988009 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 10 |
Hb_004586_120 |
0.1039722153 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 11 |
Hb_002252_030 |
0.1045151694 |
- |
- |
hypothetical protein JCGZ_06771 [Jatropha curcas] |
| 12 |
Hb_009707_010 |
0.1051864048 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 13 |
Hb_006648_030 |
0.1079807933 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.3, putative [Ricinus communis] |
| 14 |
Hb_078171_010 |
0.1081632841 |
- |
- |
PREDICTED: uncharacterized RING finger protein P8B7.15c-like [Jatropha curcas] |
| 15 |
Hb_106810_010 |
0.1173501736 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 16 |
Hb_034581_010 |
0.1204210452 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 17 |
Hb_000338_020 |
0.1246526631 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 18 |
Hb_000359_230 |
0.1252094544 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 19 |
Hb_004032_220 |
0.1253280007 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 20 |
Hb_002548_030 |
0.1266714483 |
- |
- |
PREDICTED: uncharacterized protein LOC103343650 [Prunus mume] |