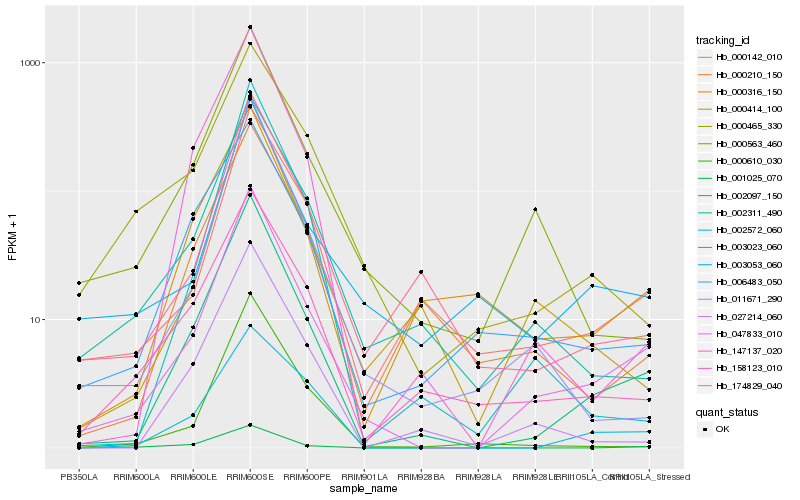
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_047833_010 |
0.0 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 2 |
Hb_002097_150 |
0.0978905987 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 3 |
Hb_000142_010 |
0.1539319883 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 4 |
Hb_000563_460 |
0.1607259316 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 5 |
Hb_002311_490 |
0.1613383085 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 6 |
Hb_003053_060 |
0.1622464728 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 7 |
Hb_011671_290 |
0.163749586 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 8 |
Hb_158123_010 |
0.1671935378 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_001025_070 |
0.1672754382 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 10 |
Hb_000414_100 |
0.1680727542 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 11 |
Hb_174829_040 |
0.1698868401 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 12 |
Hb_000610_030 |
0.1741174868 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |
| 13 |
Hb_027214_060 |
0.1766815333 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 14 |
Hb_006483_050 |
0.177756269 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 15 |
Hb_003023_060 |
0.1793573281 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 16 |
Hb_000316_150 |
0.1802027072 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_000465_330 |
0.1807680414 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 18 |
Hb_002572_060 |
0.1838795757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_147137_020 |
0.1846156757 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 20 |
Hb_000210_150 |
0.1877265548 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |