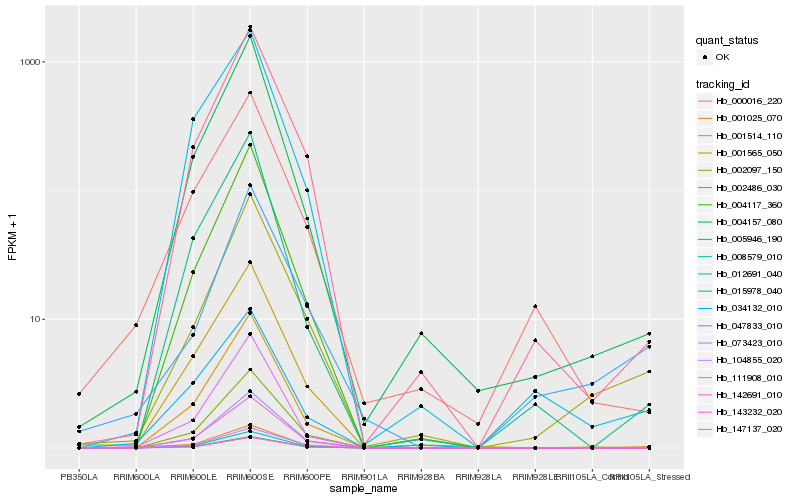
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001025_070 |
0.0 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 2 |
Hb_002097_150 |
0.1459852514 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 3 |
Hb_147137_020 |
0.1655161478 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 4 |
Hb_047833_010 |
0.1672754382 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 5 |
Hb_034132_010 |
0.1719917875 |
- |
- |
hypothetical protein B456_007G015400 [Gossypium raimondii] |
| 6 |
Hb_004157_080 |
0.1734833242 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |
| 7 |
Hb_001565_050 |
0.1747455091 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Eucalyptus grandis] |
| 8 |
Hb_142691_010 |
0.175330793 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 9 |
Hb_000016_220 |
0.1759450721 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 10 |
Hb_008579_010 |
0.1761087515 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 11 |
Hb_012691_040 |
0.1764185805 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Fragaria vesca subsp. vesca] |
| 12 |
Hb_002486_030 |
0.1769242787 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 13 |
Hb_073423_010 |
0.1776912781 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_111908_010 |
0.1802285459 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_001514_110 |
0.1803361646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004117_360 |
0.1830899708 |
- |
- |
hypothetical protein JCGZ_00367 [Jatropha curcas] |
| 17 |
Hb_104855_020 |
0.1839964813 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_143232_020 |
0.1848937451 |
- |
- |
hypothetical protein JCGZ_20467 [Jatropha curcas] |
| 19 |
Hb_005946_190 |
0.1850432953 |
- |
- |
hypothetical protein EUGRSUZ_C01051 [Eucalyptus grandis] |
| 20 |
Hb_015978_040 |
0.1880330825 |
- |
- |
PREDICTED: uncharacterized protein LOC105642609 [Jatropha curcas] |