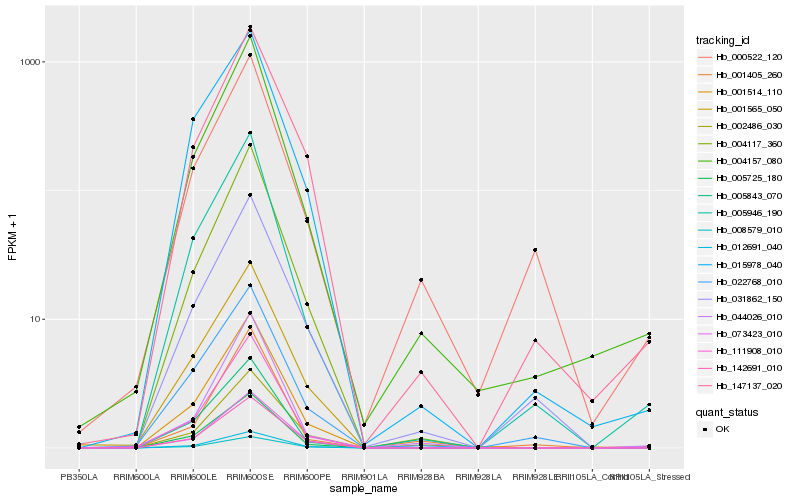
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004157_080 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |
| 2 |
Hb_005946_190 |
0.0737565676 |
- |
- |
hypothetical protein EUGRSUZ_C01051 [Eucalyptus grandis] |
| 3 |
Hb_004117_360 |
0.083187907 |
- |
- |
hypothetical protein JCGZ_00367 [Jatropha curcas] |
| 4 |
Hb_111908_010 |
0.0841438539 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_001514_110 |
0.0861947252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005725_180 |
0.0875983928 |
- |
- |
- |
| 7 |
Hb_044026_010 |
0.0904941382 |
- |
- |
PREDICTED: cytochrome P450 93A3-like [Jatropha curcas] |
| 8 |
Hb_005843_070 |
0.0930294965 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 9 |
Hb_001565_050 |
0.0936456348 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Eucalyptus grandis] |
| 10 |
Hb_012691_040 |
0.0956649214 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Fragaria vesca subsp. vesca] |
| 11 |
Hb_147137_020 |
0.0990886177 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 12 |
Hb_015978_040 |
0.1041446315 |
- |
- |
PREDICTED: uncharacterized protein LOC105642609 [Jatropha curcas] |
| 13 |
Hb_022768_010 |
0.1042353574 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 14 |
Hb_073423_010 |
0.106305061 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_142691_010 |
0.1087452964 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 16 |
Hb_002486_030 |
0.1104225713 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 17 |
Hb_008579_010 |
0.1113516806 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 18 |
Hb_031862_150 |
0.1116436782 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 19 |
Hb_001405_260 |
0.1118137034 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 20 |
Hb_000522_120 |
0.1121186649 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |