| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
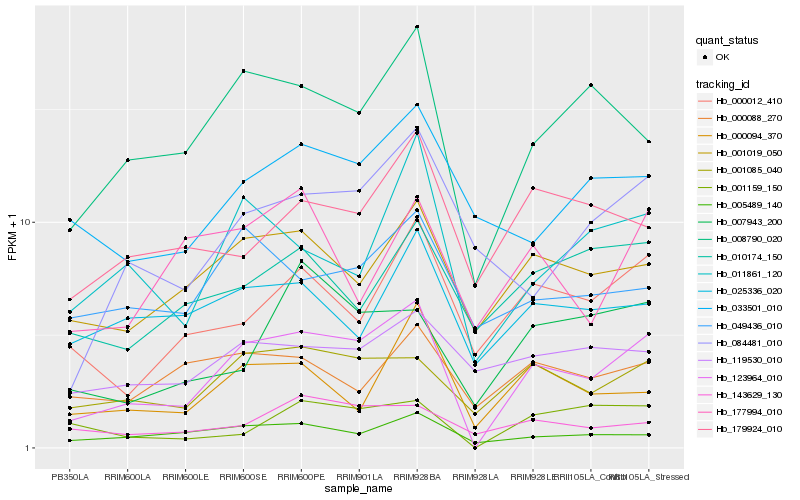
Hb_123964_010 |
0.0 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 2 |
Hb_008790_020 |
0.1769681892 |
- |
- |
PREDICTED: uncharacterized protein LOC105638122 [Jatropha curcas] |
| 3 |
Hb_001085_040 |
0.1845402534 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 4 |
Hb_001019_050 |
0.1957024331 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 5 |
Hb_011861_120 |
0.1986213932 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 6 |
Hb_005489_140 |
0.2017987524 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 7 |
Hb_001159_150 |
0.2091389648 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000012_410 |
0.2094888623 |
- |
- |
PREDICTED: histone deacetylase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000094_370 |
0.214305166 |
- |
- |
- |
| 10 |
Hb_084481_010 |
0.2171459949 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 11 |
Hb_007943_200 |
0.219403423 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 12 |
Hb_143629_130 |
0.2202384191 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_049436_010 |
0.2223960709 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 14 |
Hb_025336_020 |
0.2237136386 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 15 |
Hb_179924_010 |
0.2245994662 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_033501_010 |
0.2248971066 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 17 |
Hb_010174_150 |
0.2258728014 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_119530_010 |
0.2272329477 |
- |
- |
- |
| 19 |
Hb_000088_270 |
0.2287913157 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 20 |
Hb_177994_010 |
0.230614215 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |