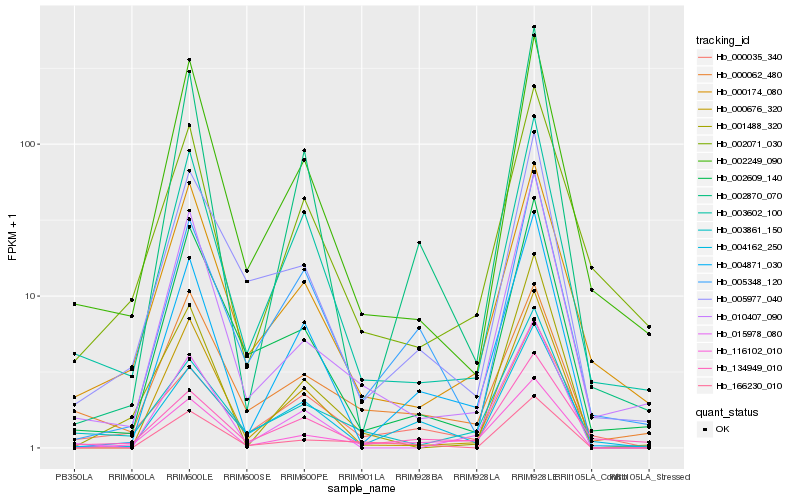
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116102_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0204s00240g [Populus trichocarpa] |
| 2 |
Hb_004871_030 |
0.1419159139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003861_150 |
0.1599754626 |
- |
- |
hypothetical protein JCGZ_15167 [Jatropha curcas] |
| 4 |
Hb_010407_090 |
0.1647016974 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 5 |
Hb_166230_010 |
0.1675099821 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_000174_080 |
0.1690623808 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 7 |
Hb_005977_040 |
0.1713469101 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 8 |
Hb_000676_320 |
0.1782253727 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 9 |
Hb_002870_070 |
0.1802743828 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 10 |
Hb_003602_100 |
0.1803755967 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_002249_090 |
0.1811968664 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004162_250 |
0.1818813273 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 13 |
Hb_002609_140 |
0.1832861422 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_005348_120 |
0.1841616453 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001488_320 |
0.1857382113 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_015978_080 |
0.1859146314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000062_480 |
0.1863524304 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002071_030 |
0.1863602109 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |
| 19 |
Hb_000035_340 |
0.1873119999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_134949_010 |
0.1879214341 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |