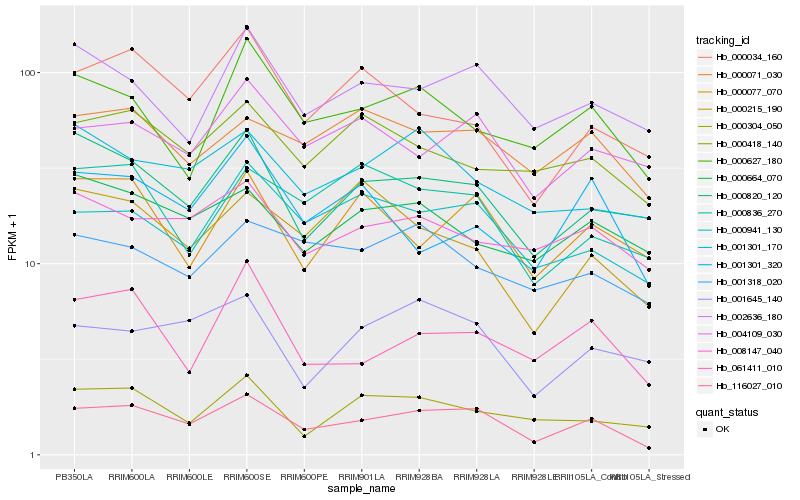
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116027_010 |
0.0 |
- |
- |
PREDICTED: separase [Jatropha curcas] |
| 2 |
Hb_000941_130 |
0.1292673051 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 3 |
Hb_000820_120 |
0.1356803639 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 4 |
Hb_002636_180 |
0.139222527 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 5 |
Hb_000071_030 |
0.142670852 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT31 [Jatropha curcas] |
| 6 |
Hb_000627_180 |
0.143491389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_061411_010 |
0.1436817391 |
- |
- |
PREDICTED: ATPase WRNIP1 [Jatropha curcas] |
| 8 |
Hb_001645_140 |
0.1452909511 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 9 |
Hb_000836_270 |
0.1464437102 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 10 |
Hb_004109_030 |
0.1467343732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000077_070 |
0.1487407547 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 12 |
Hb_008147_040 |
0.1487872234 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 13 |
Hb_000034_160 |
0.1509021601 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 14 |
Hb_001301_170 |
0.1511095871 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 15 |
Hb_000418_140 |
0.1524974493 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 16 |
Hb_001301_320 |
0.1543774953 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 17 |
Hb_000664_070 |
0.1543942222 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 18 |
Hb_001318_020 |
0.1544803143 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_000304_050 |
0.1549662794 |
transcription factor |
TF Family: GRAS |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 20 |
Hb_000215_190 |
0.1549785066 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |