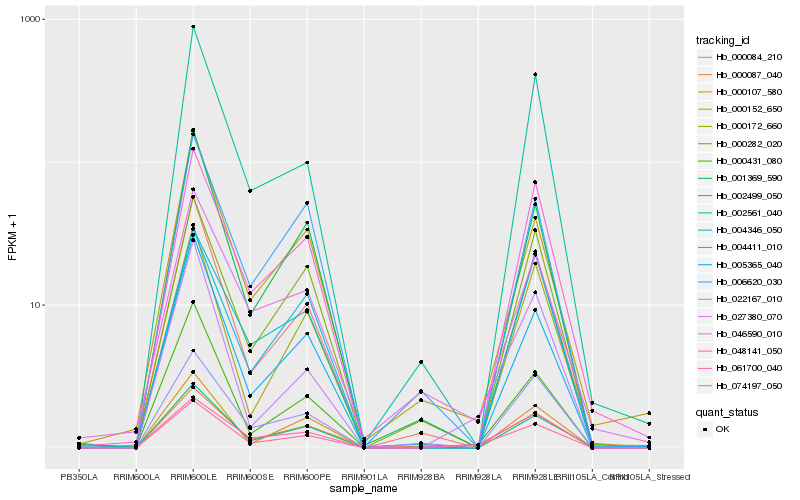
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074197_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 2 |
Hb_061700_040 |
0.1000826725 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 3 |
Hb_000084_210 |
0.1273687832 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 4 |
Hb_027380_070 |
0.1276374754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000107_580 |
0.1314538516 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 6 |
Hb_005365_040 |
0.1324259698 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 7 |
Hb_048141_050 |
0.132746275 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 8 |
Hb_002499_050 |
0.1328916701 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001369_590 |
0.1350613448 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 10 |
Hb_002561_040 |
0.1386660805 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 11 |
Hb_000431_080 |
0.1420973503 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 12 |
Hb_000087_040 |
0.1438675971 |
- |
- |
Autoinhibited Ca(2+)-ATPase 10 isoform 1 [Theobroma cacao] |
| 13 |
Hb_000282_020 |
0.1465237714 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000152_650 |
0.1467516882 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_046590_010 |
0.1470833426 |
- |
- |
Cycloeucalenol cycloisomerase, putative [Ricinus communis] |
| 16 |
Hb_004346_050 |
0.1479646818 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 17 |
Hb_000172_660 |
0.1489525817 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 18 |
Hb_022167_010 |
0.149234689 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 19 |
Hb_004411_010 |
0.1493938955 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |
| 20 |
Hb_006620_030 |
0.1506801778 |
- |
- |
nitrate reductase, putative [Ricinus communis] |