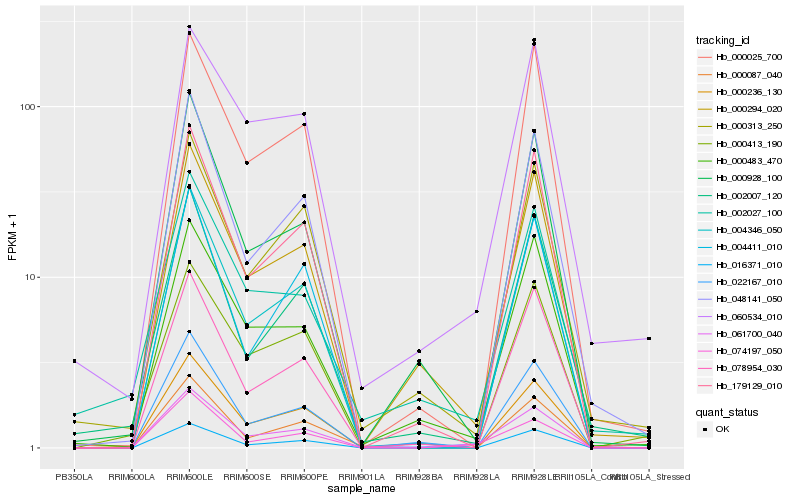
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061700_040 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 2 |
Hb_074197_050 |
0.1000826725 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 3 |
Hb_004346_050 |
0.1184440207 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 4 |
Hb_048141_050 |
0.1242858637 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 5 |
Hb_000087_040 |
0.1246402049 |
- |
- |
Autoinhibited Ca(2+)-ATPase 10 isoform 1 [Theobroma cacao] |
| 6 |
Hb_016371_010 |
0.1257649989 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 7 |
Hb_179129_010 |
0.1294242234 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 8 |
Hb_000294_020 |
0.1347618433 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 9 |
Hb_000928_100 |
0.1349127916 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 10 |
Hb_078954_030 |
0.1356240162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000483_470 |
0.137078668 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_060534_010 |
0.1373270813 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 13 |
Hb_000313_250 |
0.1382796159 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 14 |
Hb_000025_700 |
0.1393310331 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 15 |
Hb_000413_190 |
0.1417345087 |
- |
- |
PREDICTED: oxygen-evolving enhancer protein 3-2, chloroplastic [Pyrus x bretschneideri] |
| 16 |
Hb_000236_130 |
0.1417831051 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 17 |
Hb_002007_120 |
0.1420065247 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 18 |
Hb_022167_010 |
0.1420620261 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 19 |
Hb_002027_100 |
0.1422345656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004411_010 |
0.1427106994 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |