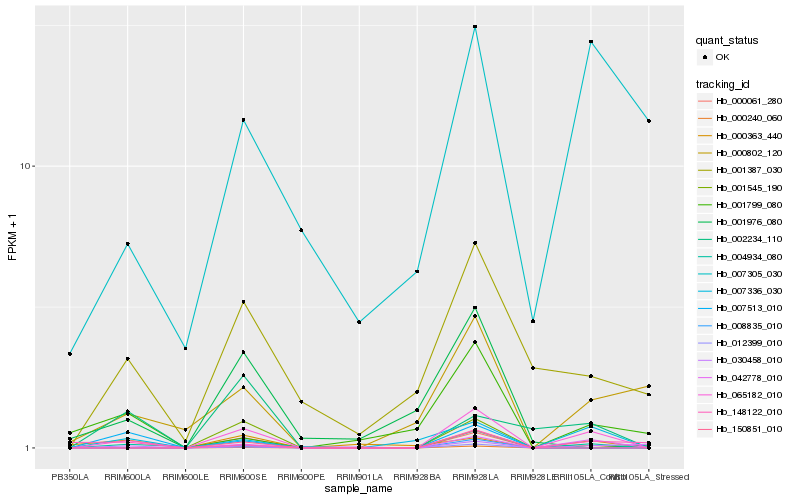
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065182_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_20878 [Jatropha curcas] |
| 2 |
Hb_042778_010 |
0.2503340089 |
- |
- |
- |
| 3 |
Hb_007336_030 |
0.2873078132 |
- |
- |
- |
| 4 |
Hb_004934_080 |
0.3149458247 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 5 |
Hb_000802_120 |
0.374232093 |
- |
- |
- |
| 6 |
Hb_001387_030 |
0.3828280645 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0013s13920g [Populus trichocarpa] |
| 7 |
Hb_000363_440 |
0.3870031144 |
- |
- |
- |
| 8 |
Hb_150851_010 |
0.3933238316 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 9 |
Hb_000061_280 |
0.3950497186 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g44230 [Jatropha curcas] |
| 10 |
Hb_008835_010 |
0.3951850428 |
- |
- |
PREDICTED: uncharacterized protein LOC105851721 [Cicer arietinum] |
| 11 |
Hb_002234_110 |
0.3961564179 |
- |
- |
PREDICTED: uncharacterized protein LOC103455142 [Malus domestica] |
| 12 |
Hb_148122_010 |
0.4003310823 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 13 |
Hb_012399_010 |
0.4026669242 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_007513_010 |
0.4028856255 |
- |
- |
- |
| 15 |
Hb_000240_060 |
0.4063710381 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 16 |
Hb_001545_190 |
0.4112069334 |
- |
- |
hypothetical protein VITISV_012031 [Vitis vinifera] |
| 17 |
Hb_007305_030 |
0.4143288316 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_030458_010 |
0.4158074193 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_001799_080 |
0.4187753272 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 20 |
Hb_001976_080 |
0.4208541749 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |