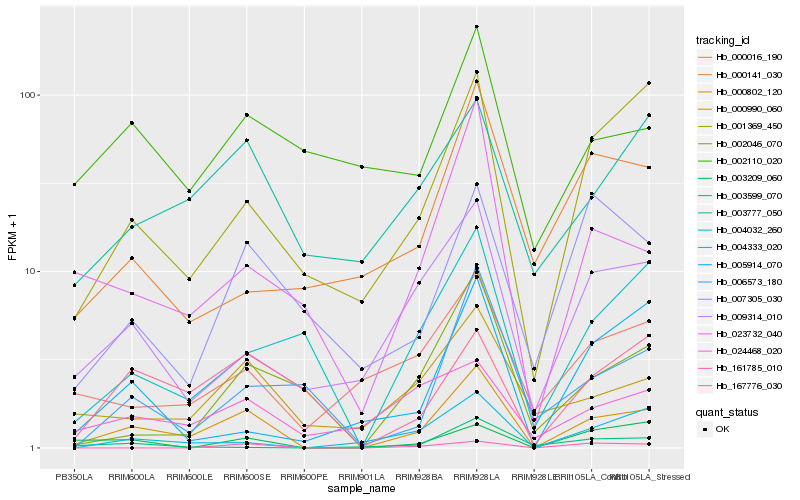
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000802_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_001369_450 |
0.2441564316 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_023732_040 |
0.2523662786 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000990_060 |
0.2574709464 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 5 |
Hb_009314_010 |
0.2585043097 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 6 |
Hb_004333_020 |
0.2643036075 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 7 |
Hb_002046_070 |
0.2671207115 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like isoform X1 [Populus euphratica] |
| 8 |
Hb_003777_050 |
0.2718364565 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 9 |
Hb_167776_030 |
0.271914031 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 10 |
Hb_004032_260 |
0.2719952004 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |
| 11 |
Hb_000016_190 |
0.2770499334 |
- |
- |
ABC transporter B family member 28 [Glycine soja] |
| 12 |
Hb_024468_020 |
0.2816629333 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 13 |
Hb_006573_180 |
0.2843335525 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 14 |
Hb_003209_060 |
0.2844437562 |
- |
- |
- |
| 15 |
Hb_007305_030 |
0.2904056035 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 16 |
Hb_161785_010 |
0.2939574722 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 17 |
Hb_003599_070 |
0.3003790658 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_005914_070 |
0.300530918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002110_020 |
0.3006373216 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 20 |
Hb_000141_030 |
0.3048471772 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |