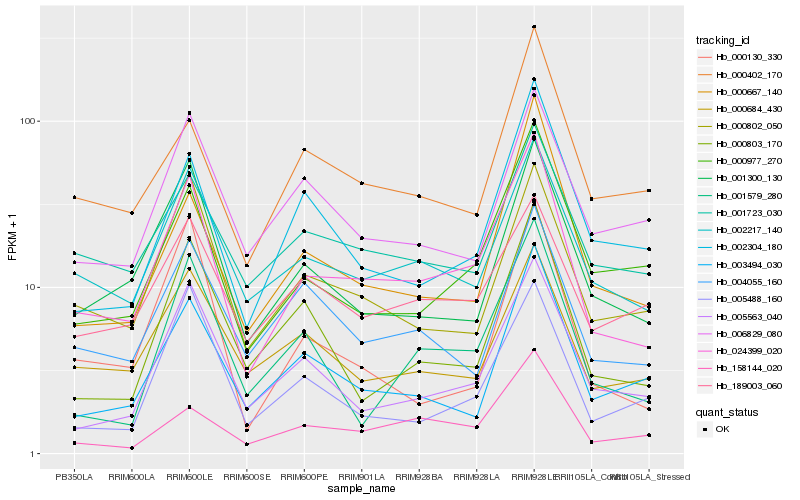
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024399_020 |
0.0 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 2 |
Hb_002217_140 |
0.1249980232 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 3 |
Hb_001300_130 |
0.1405578104 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 4 |
Hb_005563_040 |
0.142587113 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000802_050 |
0.1452832486 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 6 |
Hb_000977_270 |
0.1475684931 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000684_430 |
0.1536391664 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001579_280 |
0.1549417427 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA synthase, peroxisomal-like [Jatropha curcas] |
| 9 |
Hb_158144_020 |
0.1570525679 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 10 |
Hb_001723_030 |
0.1598392534 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004055_160 |
0.1600337929 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003494_030 |
0.1609141342 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000803_170 |
0.1620806675 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 14 |
Hb_000667_140 |
0.164424228 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002304_180 |
0.1651447985 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000402_170 |
0.1674666286 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 17 |
Hb_005488_160 |
0.1682039287 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_000130_330 |
0.1689998417 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |
| 19 |
Hb_006829_080 |
0.1694569831 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 20 |
Hb_189003_060 |
0.1711001803 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |