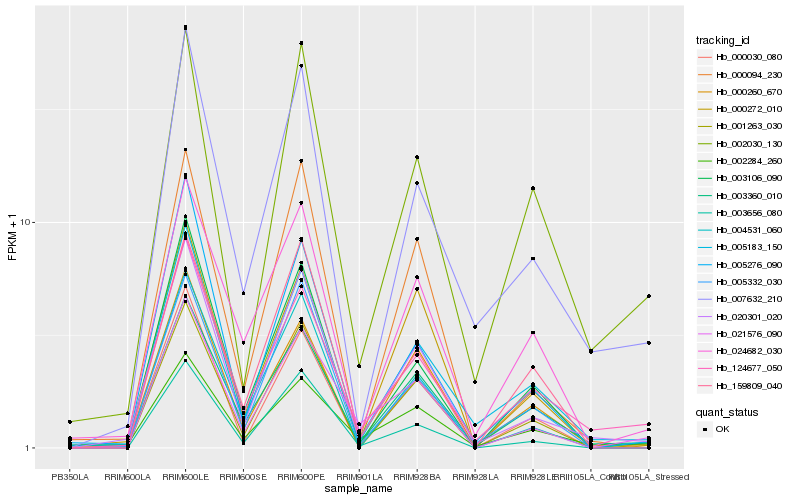
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021576_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002284_260 |
0.1367092059 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 3 |
Hb_020301_020 |
0.1383805768 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_124677_050 |
0.1392555405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003360_010 |
0.142087051 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 6 |
Hb_003656_080 |
0.1467045311 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 7 |
Hb_003106_090 |
0.155648603 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 8 |
Hb_000272_010 |
0.1624772541 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 9 |
Hb_000030_080 |
0.1628418393 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 10 |
Hb_001263_030 |
0.1631082363 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 11 |
Hb_005183_150 |
0.1658419969 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 12 |
Hb_000260_670 |
0.1661470872 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 13 |
Hb_005276_090 |
0.167196559 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004531_060 |
0.1678565906 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 15 |
Hb_024682_030 |
0.1699567332 |
- |
- |
PREDICTED: DNA replication licensing factor MCM2 [Jatropha curcas] |
| 16 |
Hb_000094_230 |
0.1704847191 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 17 |
Hb_159809_040 |
0.1719419824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005332_030 |
0.174656942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007632_210 |
0.1786442814 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 20 |
Hb_002030_130 |
0.179518799 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |