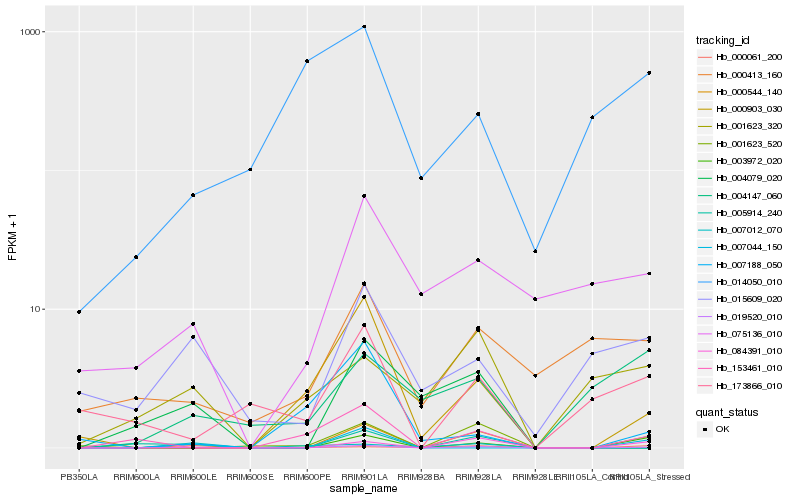
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019520_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 2 |
Hb_001623_520 |
0.3134067643 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 3 |
Hb_153461_010 |
0.3390772211 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 4 |
Hb_000903_030 |
0.3455996435 |
- |
- |
- |
| 5 |
Hb_003972_020 |
0.3640850487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_084391_010 |
0.3714747576 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 7 |
Hb_005914_240 |
0.3857543777 |
- |
- |
PREDICTED: oxidation resistance protein 1 [Jatropha curcas] |
| 8 |
Hb_001623_320 |
0.3882230311 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X1 [Populus euphratica] |
| 9 |
Hb_015609_020 |
0.3954647076 |
- |
- |
PREDICTED: uncharacterized protein LOC105642775 [Jatropha curcas] |
| 10 |
Hb_000061_200 |
0.4014153562 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 11 |
Hb_075136_010 |
0.4072290027 |
- |
- |
Protein SSU72, putative [Ricinus communis] |
| 12 |
Hb_173866_010 |
0.4099265754 |
- |
- |
- |
| 13 |
Hb_007012_070 |
0.4108322853 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 14 |
Hb_000544_140 |
0.4111853841 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 15 |
Hb_000413_160 |
0.4154794256 |
- |
- |
PREDICTED: uncharacterized protein LOC105649406 isoform X2 [Jatropha curcas] |
| 16 |
Hb_014050_010 |
0.4170974834 |
- |
- |
PREDICTED: histone H3.3-like, partial [Phoenix dactylifera] |
| 17 |
Hb_007188_050 |
0.4175837568 |
- |
- |
- |
| 18 |
Hb_007044_150 |
0.419743151 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 8A-like [Jatropha curcas] |
| 19 |
Hb_004147_060 |
0.4212069217 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 20 |
Hb_004079_020 |
0.4218426299 |
- |
- |
hypothetical protein glysoja_031528 [Glycine soja] |