| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
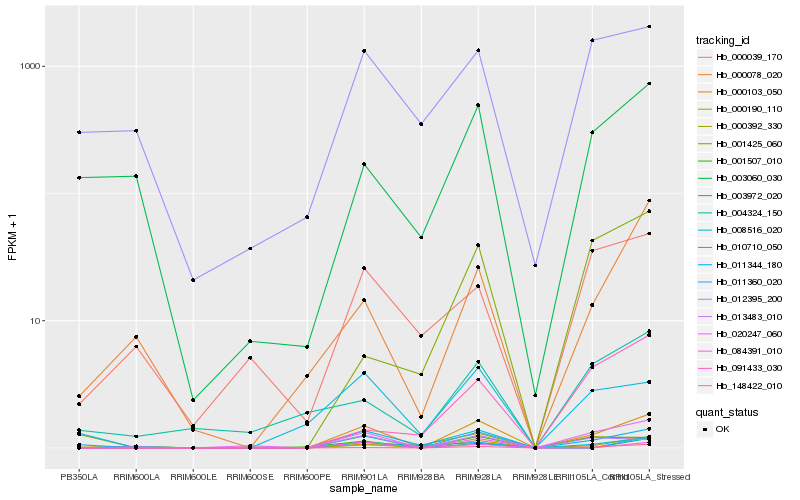
Hb_000103_050 |
0.0 |
- |
- |
hypothetical protein CICLE_v10006287mg [Citrus clementina] |
| 2 |
Hb_013483_010 |
0.2055497287 |
- |
- |
- |
| 3 |
Hb_001507_010 |
0.2060704012 |
- |
- |
PREDICTED: xylose isomerase [Jatropha curcas] |
| 4 |
Hb_010710_050 |
0.266829787 |
- |
- |
- |
| 5 |
Hb_008516_020 |
0.278776861 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 6 |
Hb_148422_010 |
0.2836228525 |
- |
- |
PREDICTED: uncharacterized protein LOC104908874, partial [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_084391_010 |
0.2880140315 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 8 |
Hb_011360_020 |
0.2914165059 |
- |
- |
- |
| 9 |
Hb_000078_020 |
0.2977872074 |
- |
- |
- |
| 10 |
Hb_011344_180 |
0.303778389 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 11 |
Hb_001425_060 |
0.307190889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000190_110 |
0.3098327022 |
- |
- |
- |
| 13 |
Hb_012395_200 |
0.3169152755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_091433_030 |
0.3182273321 |
- |
- |
- |
| 15 |
Hb_003972_020 |
0.3224883312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_020247_060 |
0.3238878981 |
- |
- |
UDP-glucoronosyl/UDP-glucosyl transferase family protein [Populus trichocarpa] |
| 17 |
Hb_004324_150 |
0.3246167579 |
- |
- |
- |
| 18 |
Hb_003060_030 |
0.324793798 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 19 |
Hb_000039_170 |
0.324879952 |
- |
- |
protein MOTHER of FT and TF 1-like [Jatropha curcas] |
| 20 |
Hb_000392_330 |
0.3252210809 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic-like [Jatropha curcas] |