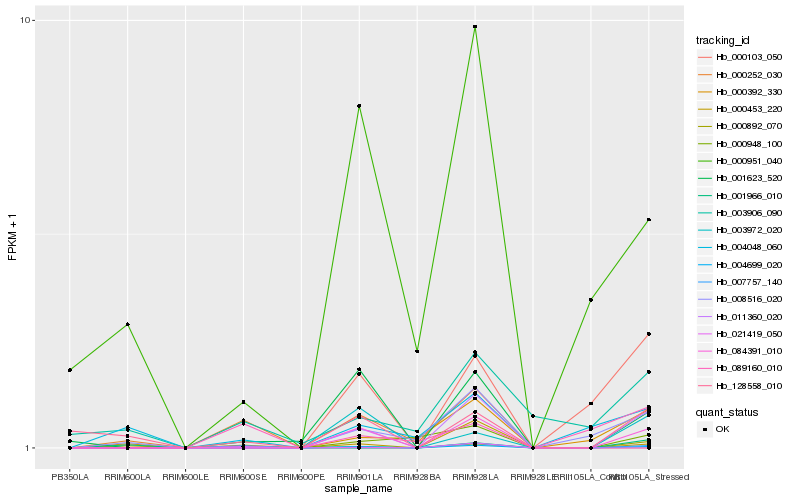
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004699_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103410170 [Malus domestica] |
| 2 |
Hb_084391_010 |
0.2576602441 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 3 |
Hb_011360_020 |
0.2730449946 |
- |
- |
- |
| 4 |
Hb_001623_520 |
0.2759613478 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 5 |
Hb_007757_140 |
0.355334672 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 6 |
Hb_128558_010 |
0.362790587 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_000951_040 |
0.365130002 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_09931 [Jatropha curcas] |
| 8 |
Hb_000392_330 |
0.3748766167 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000103_050 |
0.3831756176 |
- |
- |
hypothetical protein CICLE_v10006287mg [Citrus clementina] |
| 10 |
Hb_001966_010 |
0.3865713575 |
- |
- |
PREDICTED: uncharacterized protein LOC102611294 [Citrus sinensis] |
| 11 |
Hb_003972_020 |
0.3879393885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000453_220 |
0.3923466641 |
- |
- |
hypothetical protein JCGZ_11079 [Jatropha curcas] |
| 13 |
Hb_008516_020 |
0.3937388213 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 14 |
Hb_089160_010 |
0.3985025009 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 15 |
Hb_021419_050 |
0.4025658263 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Malus domestica] |
| 16 |
Hb_000252_030 |
0.4029673606 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 17 |
Hb_000892_070 |
0.4055132759 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004048_060 |
0.4135222432 |
- |
- |
hypothetical protein PRUPE_ppa023788mg, partial [Prunus persica] |
| 19 |
Hb_000948_100 |
0.416718077 |
- |
- |
- |
| 20 |
Hb_003906_090 |
0.4185150269 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |