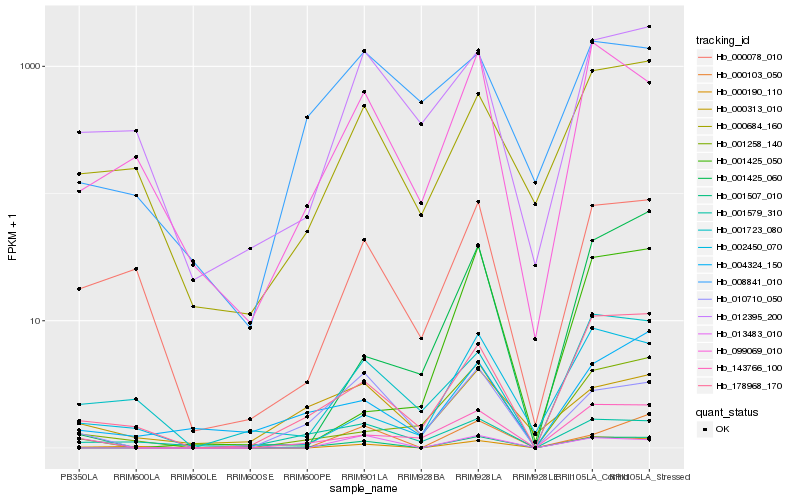
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001507_010 |
0.0 |
- |
- |
PREDICTED: xylose isomerase [Jatropha curcas] |
| 2 |
Hb_013483_010 |
0.1774617661 |
- |
- |
- |
| 3 |
Hb_000103_050 |
0.2060704012 |
- |
- |
hypothetical protein CICLE_v10006287mg [Citrus clementina] |
| 4 |
Hb_010710_050 |
0.2183304884 |
- |
- |
- |
| 5 |
Hb_178968_170 |
0.2261957874 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 homolog [Jatropha curcas] |
| 6 |
Hb_000190_110 |
0.2343326167 |
- |
- |
- |
| 7 |
Hb_099069_010 |
0.239849971 |
- |
- |
S-adenosyl-methionine-sterol-C- methyltransferase [Ricinus communis] |
| 8 |
Hb_143766_100 |
0.2555745155 |
- |
- |
PREDICTED: uncharacterized protein LOC105637800 [Jatropha curcas] |
| 9 |
Hb_002450_070 |
0.2768629941 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 10 |
Hb_004324_150 |
0.2790222382 |
- |
- |
- |
| 11 |
Hb_000078_010 |
0.2803831951 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 12 |
Hb_000684_160 |
0.2811921022 |
- |
- |
PREDICTED: lipid phosphate phosphatase gamma, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001425_060 |
0.2830535645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001258_140 |
0.2835773196 |
- |
- |
- |
| 15 |
Hb_008841_010 |
0.2885468552 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 16 |
Hb_012395_200 |
0.2901716083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001425_050 |
0.2905447558 |
- |
- |
- |
| 18 |
Hb_001723_080 |
0.2916350196 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 19 |
Hb_001579_310 |
0.2935484899 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37320 [Jatropha curcas] |
| 20 |
Hb_000313_010 |
0.2936332903 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |