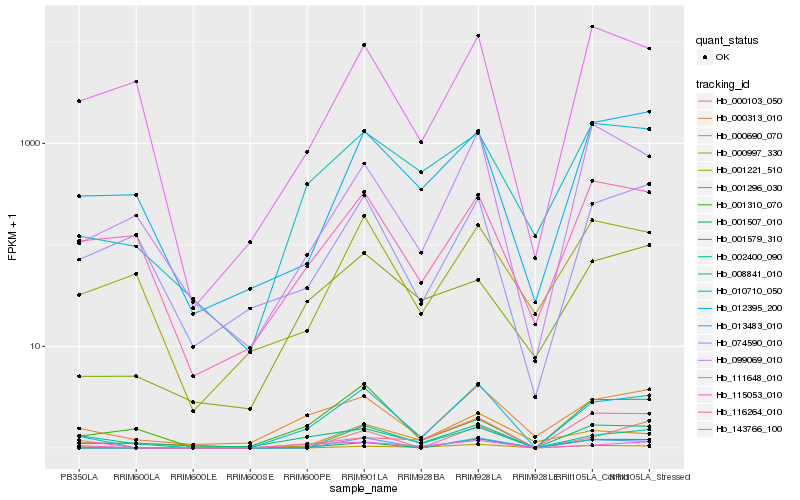
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010710_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_115053_010 |
0.1973586504 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_000313_010 |
0.1976869742 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 4 |
Hb_001579_310 |
0.2132916122 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37320 [Jatropha curcas] |
| 5 |
Hb_008841_010 |
0.2144538148 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 6 |
Hb_001507_010 |
0.2183304884 |
- |
- |
PREDICTED: xylose isomerase [Jatropha curcas] |
| 7 |
Hb_013483_010 |
0.2344448742 |
- |
- |
- |
| 8 |
Hb_000997_330 |
0.2486025861 |
- |
- |
hypothetical protein POPTR_0010s08940g [Populus trichocarpa] |
| 9 |
Hb_012395_200 |
0.2496430461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001296_030 |
0.2529218739 |
- |
- |
PREDICTED: uncharacterized protein LOC105638276 [Jatropha curcas] |
| 11 |
Hb_099069_010 |
0.2600055384 |
- |
- |
S-adenosyl-methionine-sterol-C- methyltransferase [Ricinus communis] |
| 12 |
Hb_001221_510 |
0.2601872062 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 13 |
Hb_116264_010 |
0.2615362862 |
- |
- |
hypothetical protein JCGZ_05585 [Jatropha curcas] |
| 14 |
Hb_111648_010 |
0.2631220794 |
rubber biosynthesis |
Gene Name: REF1 |
rubber elongation factor [Hevea brasiliensis] |
| 15 |
Hb_074590_010 |
0.2641506742 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 16 |
Hb_000103_050 |
0.266829787 |
- |
- |
hypothetical protein CICLE_v10006287mg [Citrus clementina] |
| 17 |
Hb_002400_090 |
0.2672034155 |
- |
- |
- |
| 18 |
Hb_000690_070 |
0.2701379353 |
- |
- |
- |
| 19 |
Hb_001310_070 |
0.2704238481 |
- |
- |
PREDICTED: uncharacterized protein LOC105639838 [Jatropha curcas] |
| 20 |
Hb_143766_100 |
0.2705524415 |
- |
- |
PREDICTED: uncharacterized protein LOC105637800 [Jatropha curcas] |