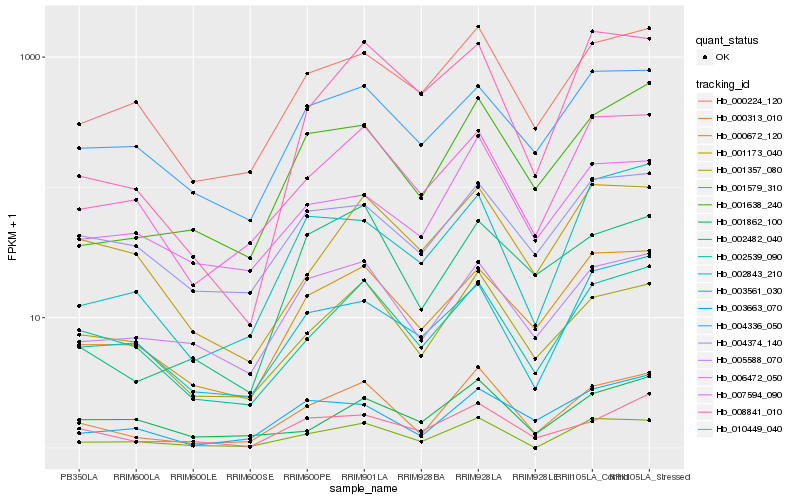
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_010 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_001638_240 |
0.1356890944 |
- |
- |
PREDICTED: uncharacterized protein LOC100262493 [Vitis vinifera] |
| 3 |
Hb_001357_080 |
0.1402540021 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 4 |
Hb_002539_090 |
0.1421528433 |
- |
- |
PREDICTED: uncharacterized protein LOC103489178 [Cucumis melo] |
| 5 |
Hb_001579_310 |
0.1446850692 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37320 [Jatropha curcas] |
| 6 |
Hb_000672_120 |
0.1480658785 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 7 |
Hb_007594_090 |
0.151050022 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18 [Jatropha curcas] |
| 8 |
Hb_000224_120 |
0.1513087907 |
- |
- |
eukaryotic translation initiation factor 1Aa [Hevea brasiliensis] |
| 9 |
Hb_010449_040 |
0.1552996626 |
- |
- |
PREDICTED: probable CDP-diacylglycerol--inositol 3-phosphatidyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_005588_070 |
0.1553489559 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 11 |
Hb_003663_070 |
0.160331665 |
- |
- |
- |
| 12 |
Hb_002482_040 |
0.1692480496 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 13 |
Hb_002843_210 |
0.1709234124 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 14 |
Hb_006472_050 |
0.1720870719 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 isoform X2 [Elaeis guineensis] |
| 15 |
Hb_003561_030 |
0.1728738756 |
- |
- |
PREDICTED: uncharacterized protein LOC105645274 [Jatropha curcas] |
| 16 |
Hb_008841_010 |
0.1740026079 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 17 |
Hb_004336_050 |
0.1754935938 |
- |
- |
hypothetical protein JCGZ_26651 [Jatropha curcas] |
| 18 |
Hb_001173_040 |
0.1811852341 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 19 |
Hb_001862_100 |
0.1822781212 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004374_140 |
0.18284154 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |