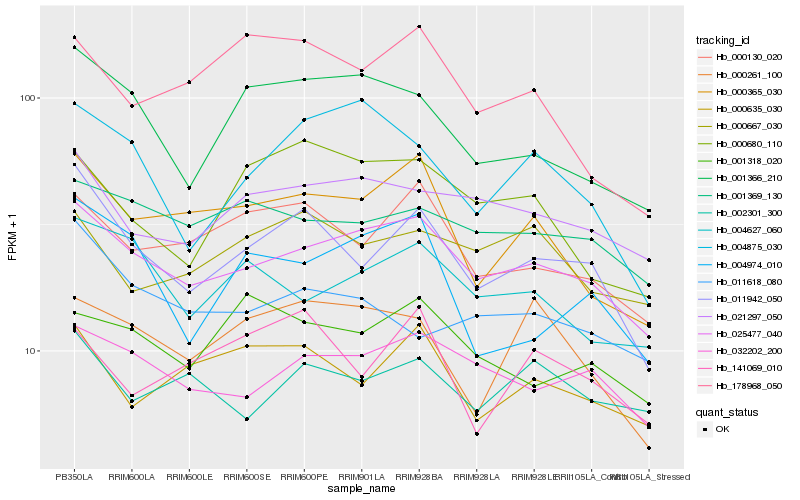
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011942_050 |
0.0 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 2 |
Hb_025477_040 |
0.0948924312 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 3 |
Hb_000130_020 |
0.1002284888 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 4 |
Hb_000365_030 |
0.1118307525 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 5 |
Hb_000635_030 |
0.1138354925 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 6 |
Hb_001366_210 |
0.115790841 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 7 |
Hb_032202_200 |
0.1157984811 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_021297_050 |
0.1226522841 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |
| 9 |
Hb_001369_130 |
0.1226573746 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011618_080 |
0.1260348678 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 11 |
Hb_141069_010 |
0.1265679254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004627_060 |
0.1268499931 |
- |
- |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 13 |
Hb_004974_010 |
0.1271421659 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 14 |
Hb_000261_100 |
0.1292166903 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000667_030 |
0.1316217086 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 16 |
Hb_001318_020 |
0.1323270071 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_000680_110 |
0.1330524011 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 18 |
Hb_002301_300 |
0.1332242855 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 19 |
Hb_004875_030 |
0.1335028718 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 20 |
Hb_178968_050 |
0.1340347477 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |