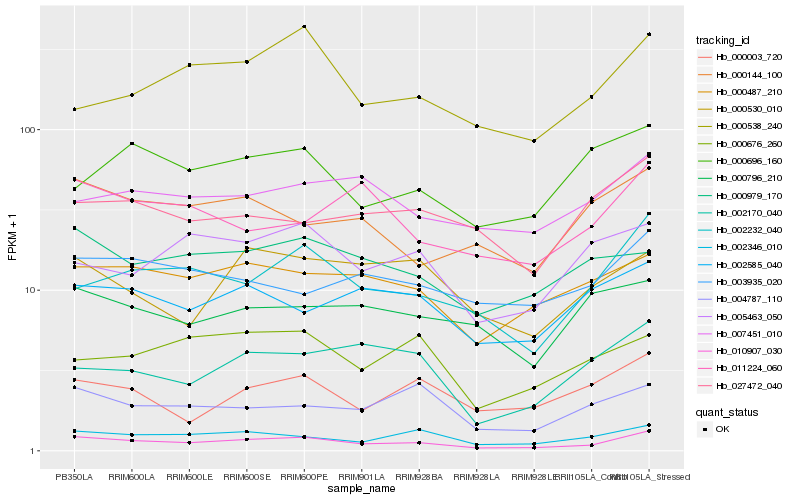
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010907_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_002232_040 |
0.1262233584 |
- |
- |
- |
| 3 |
Hb_002346_010 |
0.1293991587 |
- |
- |
- |
| 4 |
Hb_000487_210 |
0.1378297776 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 5 |
Hb_004787_110 |
0.1499867158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002585_040 |
0.1511917248 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 7 |
Hb_000003_720 |
0.1520010379 |
- |
- |
hypothetical protein POPTR_0003s04720g [Populus trichocarpa] |
| 8 |
Hb_003935_020 |
0.1520164086 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000144_100 |
0.1533332006 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002170_040 |
0.153476574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000979_170 |
0.1539339784 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 12 |
Hb_007451_010 |
0.1542374115 |
- |
- |
PREDICTED: 60S ribosome subunit biogenesis protein NIP7 homolog [Jatropha curcas] |
| 13 |
Hb_000538_240 |
0.1558436774 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 14 |
Hb_005463_050 |
0.156785991 |
- |
- |
PREDICTED: uncharacterized protein LOC105632514 [Jatropha curcas] |
| 15 |
Hb_027472_040 |
0.1575562832 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 16 |
Hb_000530_010 |
0.1577865595 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |
| 17 |
Hb_011224_060 |
0.1579634775 |
- |
- |
hypothetical protein POPTR_0011s16880g [Populus trichocarpa] |
| 18 |
Hb_000676_260 |
0.1583937109 |
- |
- |
PREDICTED: uncharacterized protein LOC105639070 [Jatropha curcas] |
| 19 |
Hb_000696_160 |
0.1597456516 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 20 |
Hb_000796_210 |
0.1601199969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |