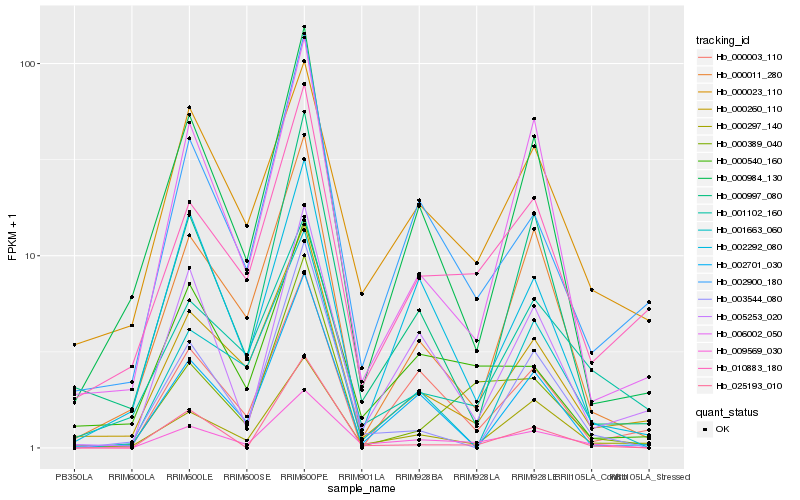
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009569_030 |
0.0 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 2 |
Hb_000297_140 |
0.1669435458 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 3 |
Hb_005253_020 |
0.1766231378 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 4 |
Hb_002900_180 |
0.1787915468 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 5 |
Hb_010883_180 |
0.1844795639 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 6 |
Hb_000011_280 |
0.1851461891 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 7 |
Hb_000260_110 |
0.1870630102 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 8 |
Hb_000984_130 |
0.1872915072 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 9 |
Hb_006002_050 |
0.1886016728 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 10 |
Hb_000997_080 |
0.1887730492 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000540_160 |
0.1900731946 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF6 [Jatropha curcas] |
| 12 |
Hb_000023_110 |
0.1904169055 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 13 |
Hb_000389_040 |
0.1909960062 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g03250-like [Jatropha curcas] |
| 14 |
Hb_002292_080 |
0.1922078442 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 15 |
Hb_000003_110 |
0.1953955667 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_003544_080 |
0.1962460017 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 17 |
Hb_002701_030 |
0.1964619259 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 18 |
Hb_001102_160 |
0.1979392079 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 19 |
Hb_001663_060 |
0.1982135616 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 20 |
Hb_025193_010 |
0.1988665637 |
- |
- |
PREDICTED: indole-3-acetic acid-amido synthetase GH3.17-like isoform X1 [Jatropha curcas] |